Figures & data
Table 1 Primer sequence information used in qRT-PCR
Table 2 Results of differential expression of LINC00882
Figure 1 High expression of LINC00882 is correlated with poor overall survival of HCC patients.
Abbreviations: HCC, hepatocellular carcinoma; GEO, Gene Expression Omnibus; Con, control.
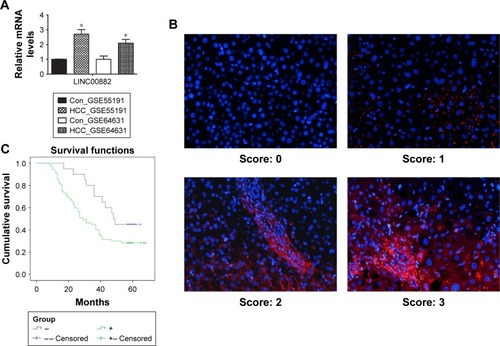
Table 3 The expression information of clinical tissues
Table 4 Association between LINC00882 expression and clinicopathological characteristics in HCC patient
Table 5 Mean and median for survival time
Table 6 Overall comparisons
Table 7 COX regression analysis
Figure 2 LINC00882 knockdown results in expression inhibition of proliferative markers and invasion-related proteins.
Abbreviations: HCC, hepatocellular carcinoma; Lv, lentivirus; MMP, matrix metalloproteinase; NC, negative control; qRT-PCR, quantitative real-time polymerase chain reaction; Con, control.
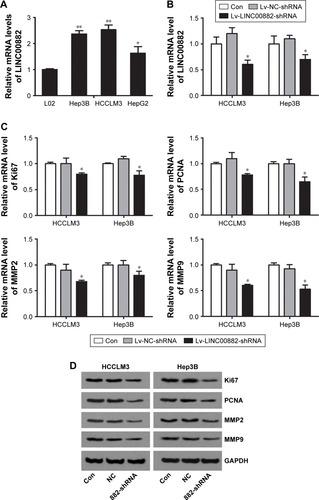
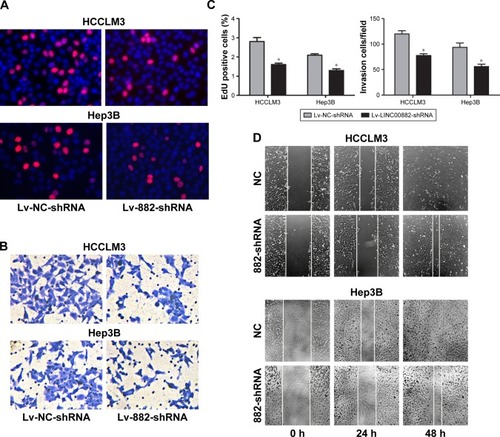
Figure 3 LINC00882 knockdown reduces abilities of proliferation, migration and invasion of HCC cells.
Abbreviations: EDU, 5-ethynyl-2′-deoxyuridine; HCC, hepatocellular carcinoma; Lv, lentivirus; NC, negative control.