Figures & data
Figure 1 Map of the most recurrent amino acid substitutions in the BCR-ABL1 kinase domain in Ph+ clinical samples.
Abbreviations: IM, imatinib; Ph+, Philadelphia chromosome positive.
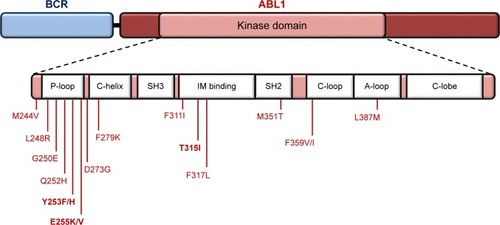
Figure 2 Cytogenetic (A) and molecular (B) analyses of Ph+ ALL patient’s bone marrow sample.
Abbreviations: ALL, acute lymphoblastic leukemia; FISH, fluorescence in situ hybridization; NTC, non-template control; Ph+, Philadelphia chromosome positive.
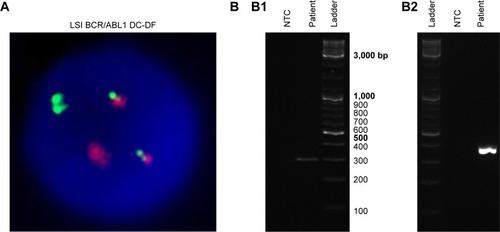
Figure 3 Ph+ ALL patient’s novel BCR-ABL1 point mutation and disease molecular monitoring.
Abbreviations: ALL, acute lymphoblastic leukemia; IM, imatinib; Ph+, Philadelphia chromosome positive; WT, wild-type.
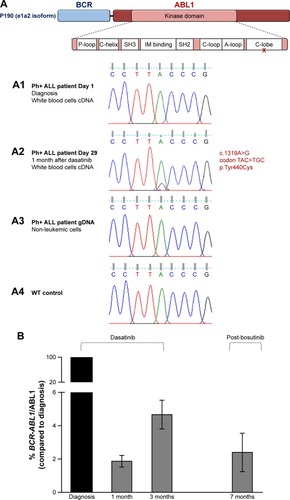
Figure S1 Alignment of Ph+ ALL patient e1a2 nucleotide sequence (Seq 1) with BCR-ABL1 e1a2 reference sequence from GenBank AF113911.1 (Seq 2).
Abbreviations: ALL, acute lymphoblastic leukemia; Ph+, Philadelphia chromosome positive.
Figure S2 DISOclust disorder prediction results.
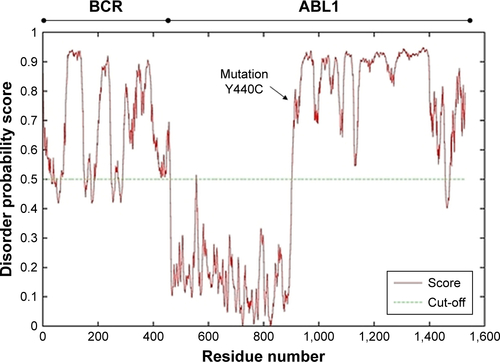
Table S1 Nested-PCR primers for the amplification of ABL1, BCR-ABL1 transcript identification and analysis of BCR-ABL1 mutational status
Table S2 ABL1 protein alignment between different species demonstrates the conservation of the affected amino acid
