Figures & data
Table 1 Specific Hp proteins that are associated with metabolic reprogramming in GC
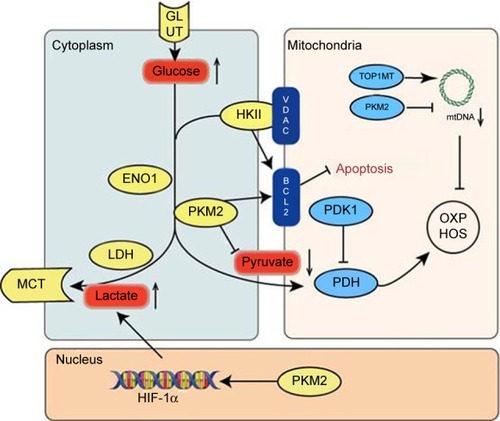
Figure 1 An overview of the pathways mediating upregulation of glycolysis and mitochondrial dysfunction in GC.
Abbreviations: GC, gastric cancer; mtDNA, mitochondrial DNA; OXPHOS, oxidative phosphorylation; PKM2, pyruvate kinase M2.
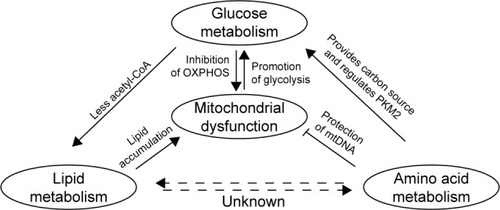
Figure 2 Metabolic reprogramming in GC.
Notes: A model is proposed in which an increase in glycolysis promotes mitochondrial dysfunction. The latter leads to disturbances in the metabolism of lipids and amino acids, and these alterations correlate with changes in the metabolism of glucose.
Abbreviations: ENO1, Enolase; GC, gastric cancer; GLUT, glucose transporter; HIF-1α, hypoxia-inducible factor 1α; HKII, hexokinase II; MCT, monocarboxylic acid transporter; LDH, lactate dehydrogenase; OXPHOS, oxidative phosphorylation; PDH, pyruvate dehydrogenase; PDK, pyruvate dehydrogenase kinase; PKM2, pyruvate kinase M2; TOP1MT, mitochondrial topoisomerase I; VDACs, voltage-dependent anion channels.