Figures & data
Figure 1 CDR1as expression in ovarian cancer tissues and cells. (A) The relative expression of CDR1as in tissues from 65 ovarian cancer patients and 37 patients without ovarian cancer was measured by qRT-PCR. (B) and (C) The relative expression of CDR1as in ovarian cancer cells (HO8910 and A2780) was analyzed by qRT-PCR. (D) CDR1as was overexpressed in ovarian cancer cells via a CDR1as-specific adenovirus (Ad-CDR1as), and its expression was detected by qRT-PCR. (E) CDR1as expression was silenced in ovarian cancer cells with a CDR1as-specific siRNA (Si-CDR1as) and then measured by qRT-PCR. All values are expressed as the mean ± SEM (*P<0.05 and ***P<0.001).
Figure 2 CDR1as inhibits the proliferation of ovarian cancer cells. (A) PCNA protein expression levels were analyzed by Western blot. (B) Ki67 expression was estimated in ovarian cancer cells after staining for Ki67; the nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Magnification, ×400. (C) The proliferation of ovarian cancer cells was assessed by MTT assay. (D) The proliferation rate of ovarian cancer cells was measured by CCK8 assay. All values are expressed as the mean ± SEM (*P<0.05, **P<0.01, and ***P<0.001).
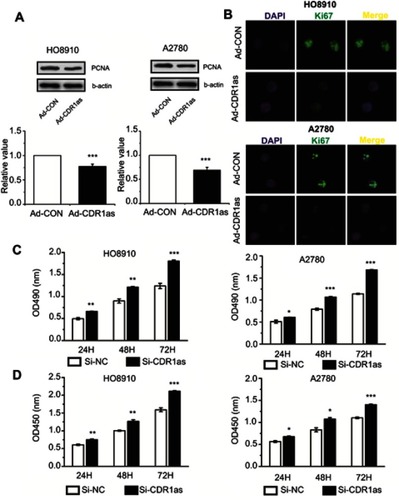
Figure 3 CDR1as inhibits the invasion and migration of ovarian cancer cells. (A) and (B) The invasion ability of ovarian cancer cells was measured by Transwell® assay. Magnification, ×100. (C) and (D) The migration ability of ovarian cancer cells was detected by scratch-wound assay. Magnification, ×100. All values are expressed as the mean ± SEM (**P<0.01 and ***P<0.001).
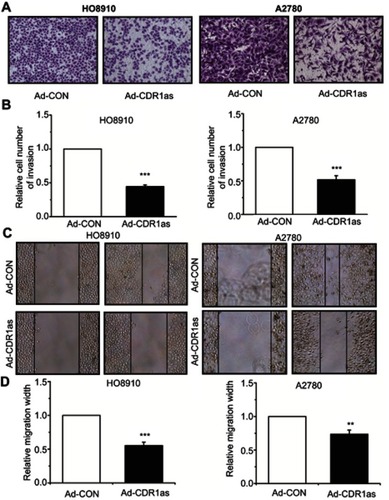
Figure 4 MiR-135b-5p promotes the proliferation of ovarian cancer cells. (A) The interaction between CDR1as and miR-135b-5p was predicted by a bioinformatics database. (B) The miR-135b-5p expression level was analyzed by qRT-PCR. (C) The proliferation of ovarian cancer cells was measured by MTT assay. (D) The proliferation rate of ovarian cancer cells was detected by CCK8 assay. All values are expressed as the mean ± SEM (*P<0.05, **P<0.01, and ***P<0.001).
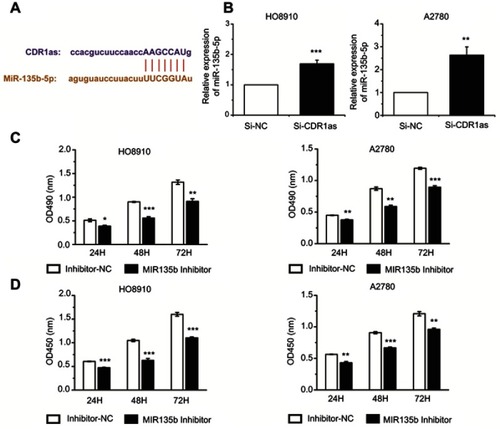
Figure 5 HIF1AN expression was positively correlated with CDR1as and negatively correlated with miR-135b-5p. (A) HIF1AN was identified as a potential target gene of miR-135b-5p by bioinformatics analysis (TargetScanHuman database). (B) and (C) HIF1AN mRNA expression levels were analyzed by qRT-PCR. (D) and (E) HIF1AN protein expression levels were analyzed by Western blotting. All values are expressed as the mean ± SEM (*P<0.05, **P<0.01, and ***P<0.001).
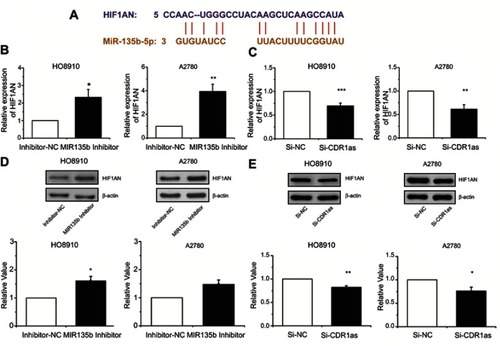
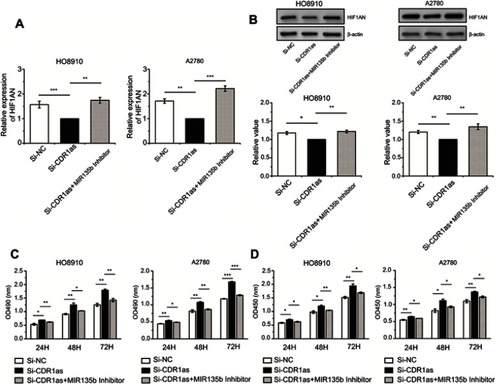
Figure 6 CDR1as targeted miR-135b-5p and promoted HIF1AN in ovarian cancer cells. (A) HIF1AN mRNA expression levels were analyzed by qRT-PCR assay. (B) HIF1AN protein expression levels were analyzed by Western blotting. (C) The proliferation of ovarian cancer cells was measured by MTT assay. (D) The proliferation ratio of ovarian cancer cells was measured by CCK8 assay. All values are expressed as the mean ± SEM (*P<0.05, **P<0.01, and ***P<0.001).