Figures & data
Table 1 Relationship between LINC00958 expression and clinicopathological variables (n=48)
Figure 1 Upregulation of LINC00958 expression in HNSCC tissues and association with poor HNSCC prognosis. (A) Analysis of GEPIA database data. The relative expression of LINC00958 in tumor vs normal tissues was compared in different types of human cancer. The height of the bar represents the median expression of tumors (red) or normal tissues (black). (B) qRT-PCR. Relative expression of LINC00958 was assessed in 48 pairs of HNSCC and normal tissues. (C-D) Kaplan-Meier curves and log-rank test. The GEPIA database (C) and Kaplan-Meier plotter website data were subjected to Kaplan-Meier curve analysis and the log-rank test (D).
Abbreviations: BLCA, bladder urothelial carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinaoma; LUSC, lung squamous cell carcinoma; OV, ovarian serous cystadenocarcinoma. T, tumor; N, normal.
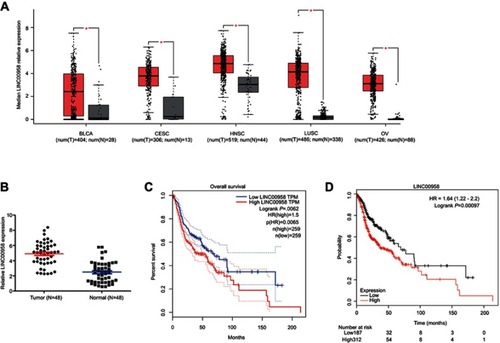
Figure 2 Induction of HNSCC cell sensitivity to ionizing radiation and cisplatin after knockdown of LINC00958 expression. (A) qRT-PCR. The relative expression of LINC00958 was assessed in HNSCC cell lines (Detroit 562, Cal27, SCC-9, SCC-15, and FaDu) and a normal oral epithelial cell line (HOK). (B) qRT-PCR. LINC00958 expression was manipulated using LINC00958 siRNAs or LINC00958 cDNA in Fadu and Detroit562 cells, respectively and assessed by using qRT-PCR. (C) CCK-8 assay. LINC00958 expression was manipulated using LINC00958 siRNAs or LINC00958 cDNA in Fadu and Detroit562 cells, respectively and assessed for changed cell viability using the CCK-8 assay. (D and F) CCK-8 assay. Fadu cells were grown and transfected with LINC00958 siRNAs and then subjected to different doses of ionizing radiation (0-10Gy) (D) or cisplatin (0–25 μM) (F) and the CCK-8 assay. (E and G) Colony formation assay. Fadu cells were grown and transfected with LINC00958 siRNAs and then subjected to 2 Gy ionizing radiation (E) or 5 µM cisplatin (G) and were subjected to colony formation assay. The data are the means ± SD (n=3). *p<0.05 and **p<0.01 by two-tailed Student’s t-test.
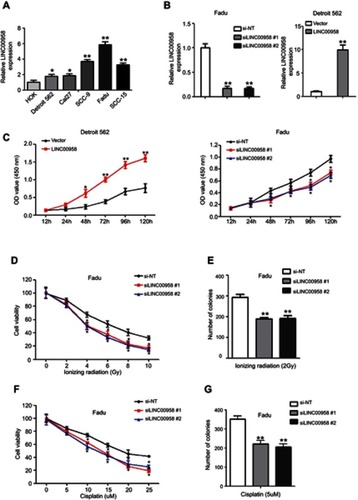
Figure 3 Suppression of c-Myc mRNA expression after knockdown of LINC00958 expression in HNSCC. (A) Bioinformatical analysis of LINC00958 and MYC expression using the starBase v3.0 Pan-Cancer Analysis and Networks Platform data. (B) qRT-PCR and Western blot. Fadu and Detroit cells were grown and subjected to c-Myc siRNA or c-Myc plasmid transfection and then qRT-PCR or Western blot analysis. (C) Bioinformatical analysis. The data showed a schematic representation of c-Myc binding site in LINC00958 promoter region. (D) ChIP-qPCR assay. HEK-293T cells were grown and subjected to immunoprecipitation with an anti-c-Myc antibody and then DNA extraction and qPCR analysis of LINC00958 promoter. (E) Illustration of the pGL3-LINC00958 promoter reporter constructs. pGL3-LINC00958 (WT) contains the wild type LINC00958 promoter region, whereas pGL3-LINC00958 (mut) contains a mutated LINC00958 promoter region to disrupt c-Myc binding. (F) Luciferase reporter assay. HEK293T cells were grown and subjected to transfection with c-Myc siRNA, luciferase reporter, LINC00958 promoter (wild type or mutated) constructs, and luciferase reporter assay. The data are means ± SD (n=3). *p<0.05 and **p<0.01 by two-tailed Student’s t-test.
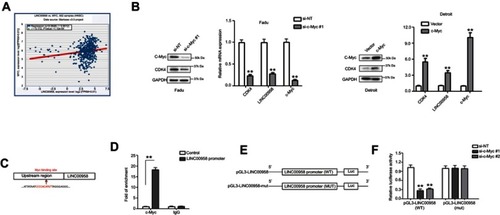
Figure 4 LINC00958 participation of c-Myc gene network. (A) The RIP assay. HEK293T cells were grown and subjected to immunoprecipitation with an anti-c-Myc antibody and then RNA isolation and qRT-PCR analysis of LINC00958 level. (B) Luciferase reporter assay. HNSCC cells were grown and subjected to transfection with c-Myc responsive luciferase reporters and LINC00958 siRNAs, nonsense siRNA, LINC00958 cDNA or vector-only and then luciferase reporter assay. (C) qRT-PCR. Fadu and Detroit562 cells were grown and transfected with LINC00958 siRNAs, nonsense siRNA, LINC00958 cDNA or vector-only, respectively and then subjected to qRT-PCR analysis of CDK2, CDC25B, CDC45, CDK4, CyclinA2, and CDC20 mRNA. (D) ChIP assay. HNSCC cells were grown and transfected with indicated siRNAs or plasmids and then subjected to immunoprecipitation with an anti-c-Myc antibody and DNA extraction and qPCR analysis. The data are means ± SD (n=3). *p<0.05 and **p<0.01 by two-tailed Student’s t-test.
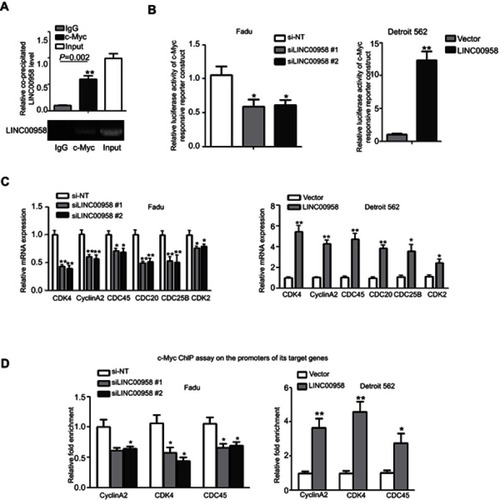
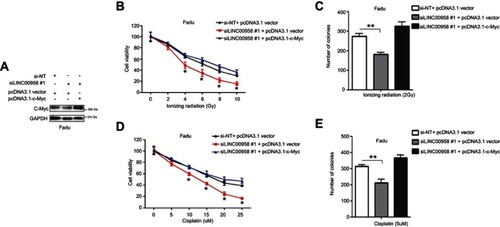
Figure 5 Regulation of HNSCC cell chemo- and radiation sensitivity by LINC00958-c-Myc positive feedback network. (A) Western blot. Fadu cells were grown and co-transfected c-Myc cDNA and siLINC00958 #1 or their negative controls and then subjected to Western blot analysis. (B and D) Cell viability CCK-8 assay. Fadu cells were grown and co-transfected c-Myc cDNA and siLINC00958 #1 or their negative controls and then subjected to different doses of ionizing radiation (0-10Gy) (B) or cisplatin (0-25μM) (D) and the CCK-8 assay. (C and E) Colony formation assay. Fadu cells were grown and co-transfected c-Myc cDNA and siLINC00958 #1 or their negative controls and then subjected to 2 Gy ionizing radiation (C) or 5 µM cisplatin (E) and the colony formation assay. The data are means ± SD (n=3). *p<0.05 and **p<0.01 by two-tailed Student’s t-test.