Figures & data
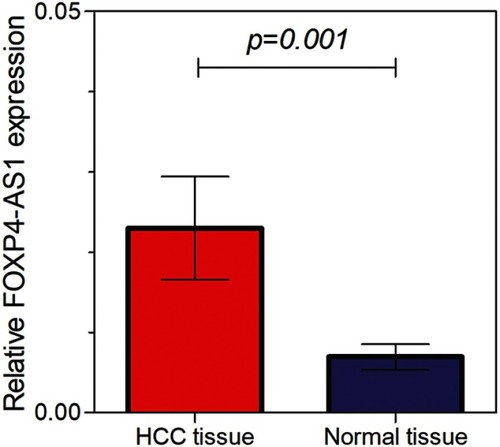
Figure 2 Kaplan-Meier curves for FOXP4-AS1 in HCC based on qRT-PCR. (A) Disease-free survivals of groups with low and high levels of FOXP4-AS1. (B) Overall survival of groups with low and high levels of FOXP4-AS1.
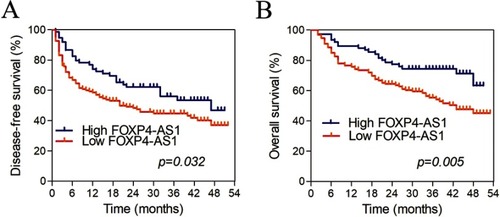
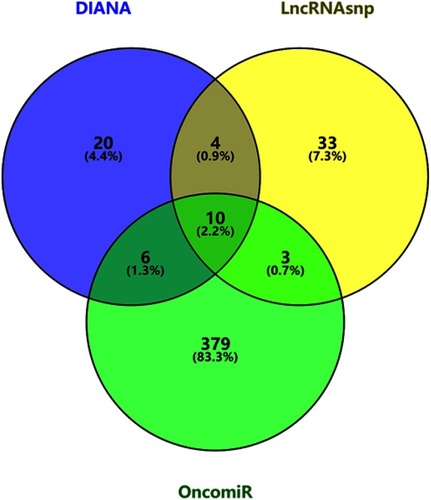
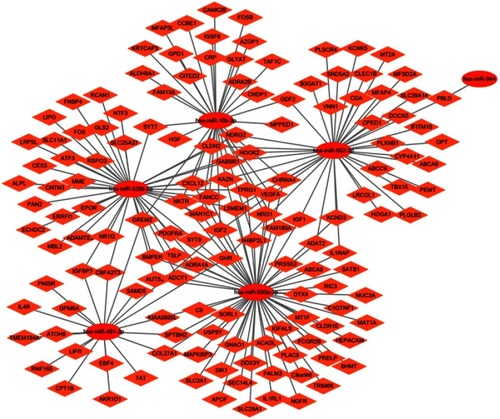
Figure 4 Venn diagram of target genes of FOXP4-AS1. (A) Upregulated target genes; (B) downregulated target genes.
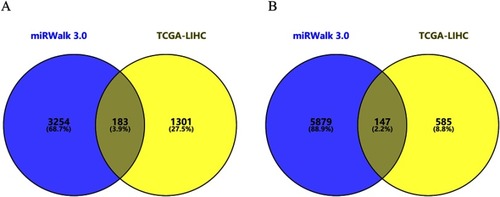
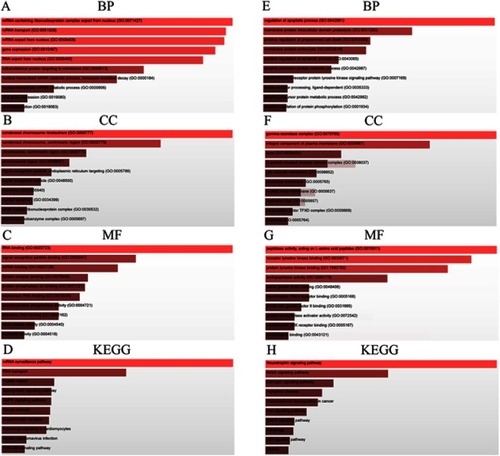
Figure 7 GO functional annotation and KEGG pathway analysis. Most prominent BP, CC, MF, and KEGG pathways associated with (A–D) upregulated and (E–H) downregulated candidate genes of FOXP4-AS1. (A, E) BP; (B, F) CC; (C, G) MF; and (D, H) KEGG pathways.
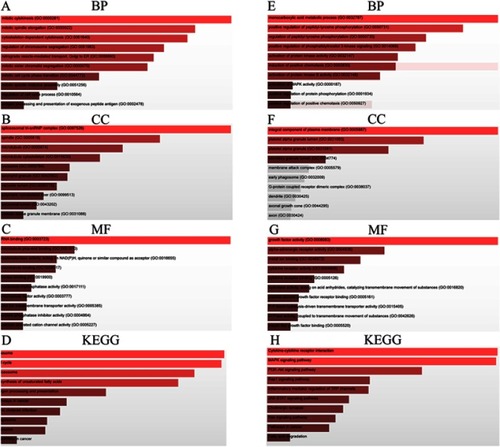
Figure 10 Expressions of 10 upregulated hub genes in HCC and normal tissues, based on GEPIA. (A) HJURP. (B) DTL. (C) CKAP2. (D) KNSTRN. (E) MCM4. (F) PRC1. (G) RPL36A. (H) SNRPD1. (I) CDCA5. (J) CENPN.
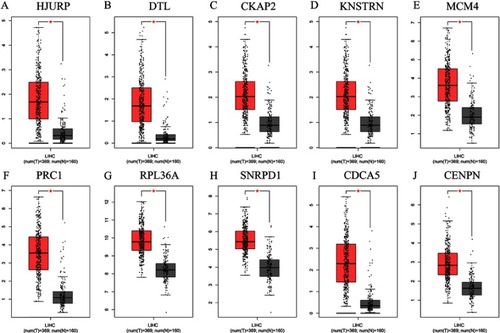
Figure 11 Expressions of 10 downregulated hub genes in HCC and normal tissues, based on GEPIA. (A) GHR. (B) FOSB. (C) ADRA2B. (D) ADRA1A. (E) SLC11A1. (F) NTF3. (G) NGFR. (H) IGFBP3. (I) CDF2. (J) GABBR1.
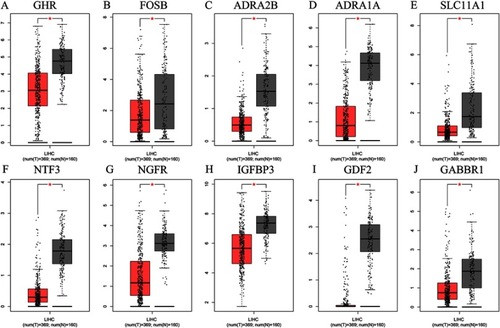
Figure 12 Association of expressions of 10 upregulated hub genes and the survival of patients with HCC. (A) CKAP2. (B) CENPN. (C) CDCA5. (D) SNRPD1. (E) RPL36A. (F) PRC1. (G) MCM4. (H) KNSTRN. (I) HJURP. (J) DTL.
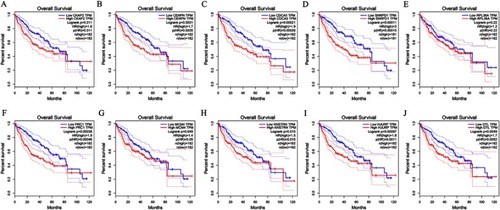
Figure 13 Association of expressions of 10 downregulated hub genes and survival of patients with HCC. (A) ADRA1A. (B) SLC11A1. (C) NTF3. (D) NGFR. (E) IGFBP3. (F) GHR. (G) GDF2. (H) GABBR1. (I) FOSB. (J) ADRA2B.
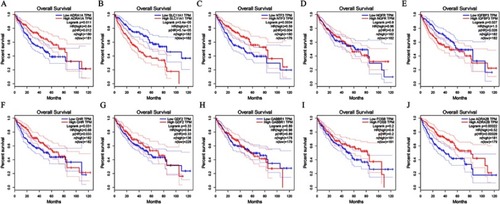
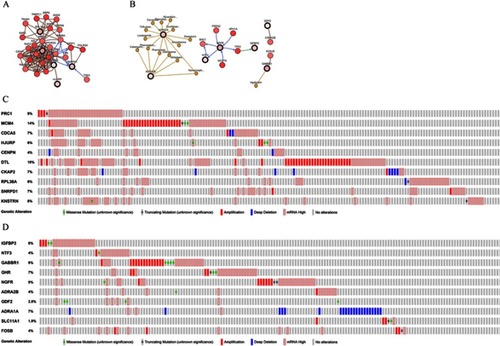
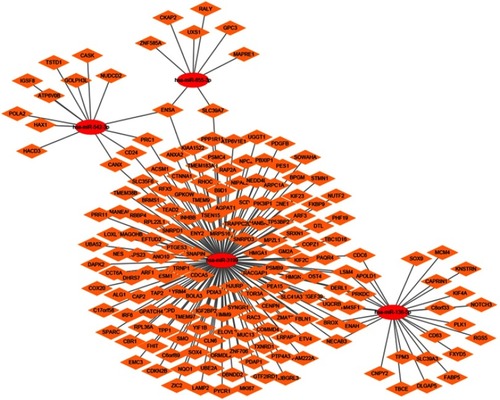
Figure 14 Network review of genes neighboring hub genes and genetic alteration of hub genes in HCC. (A) Ten upregulated hub genes. (B) Ten downregulated hub gene. (C) Genetic alteration of 10 upregulated hub genes. (D) Genetic alteration of 10 downregulated hub genes. Association between hub genes and drugs.