Figures & data
Figure 1 The distinct binding epitopes of HER2-targeted monoclonal antibodies approved by the FDA. Trastuzumab binds to subdomain IV of the HER2 extracellular domain. Pertuzumab binds to an epitope in subdomain II, the dimerization domain of HER2.
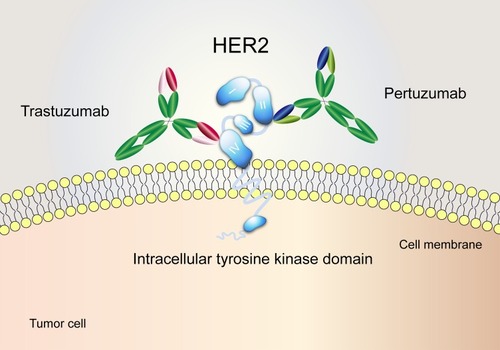
Figure 2 The distribution and frequency of HER2 S310F mutation in tumours. (A) The distribution of recurrent mutations (more than five occurrences in an individual cancer type) in the extracellular domain of membrane proteins. The numbers mean the occurrences of each mutation in the same tumour. Different cancer types are represented by different colours. (B) The distribution of HER2 mutations based on COSMIC database.
Abbreviations: BLCA, bladder urothelial carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; GBM, glioblastoma multiforme; LGG, brain lower grade glioma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; UCEC, uterine corpus endometrial carcinoma.
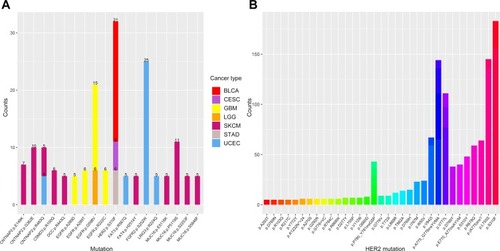
Figure 3 Molecular modelling of the interaction between pertuzumab and HER2. (A) Structure visualization of the HER2-pertuzumab complex (PDB: 1S78), including the HER2 extracellular domain (bluish-green), pertuzumab’s light chain (purple) and heavy chain (green). The position of HER2 Ser310 was confirmed at the interface of HER2 and pertuzumab. Selected side chains from HER2 and pertuzumab are shown in stick representation, with carbons coloured by domain, nitrogens blue, oxygens red and hydrogens grey. The backbones are shown in ribbon representation. (B) Molecular modelling of pertuzumab bound to WT/S310F-mutant HER2. Hydrogen bonds are shown as red dashed lines. (C) The top 10 critical residues of the HER2-pertuzumab interaction predicted by the HawkDock server.
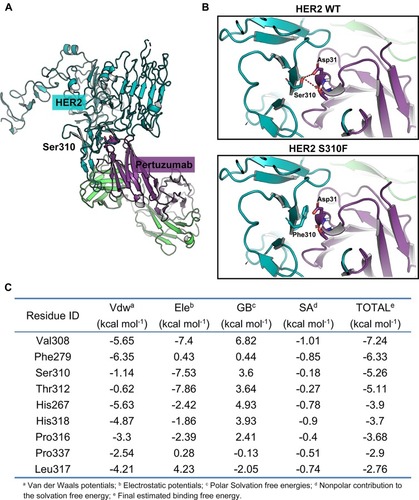
Figure 4 The binding affinities of anti-HER2 antibodies for WT/S310F-mutant HER2. (A) The binding affinities of pertuzumab and trastuzumab for NIH3T3 cells expressing WT (HER2 WT) or S310F-mutant (HER2 S310F) HER2. NIH3T3 cells were used as a negative control. (B) The affinities of serial concentrations of pertuzumab and trastuzumab for WT (NIH3T3-HER2 WT) or S310F-mutant (NIH3T3-HER2 S310F) HER2 cells. (C) Comparisons of pertuzumab and trastuzumab interacting with WT (HER2-ECD-WT) or S310F-mutant (HER2-ECD-S310F) HER2 ECD protein by ELISA as described in the materials and methods. (D) The kinetic parameters for trastuzumab and pertuzumab derived from the fitted data models according to SPR analyses.
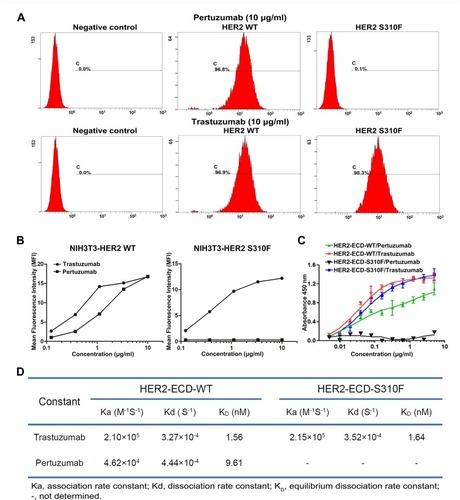
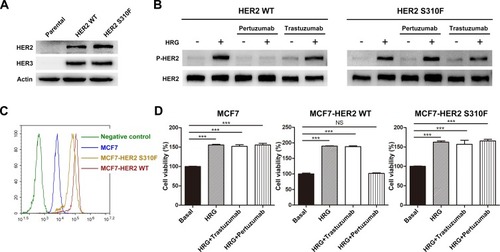
Figure 5 Effect of S310F mutation on the antitumour activity of pertuzumab. (A) NIH3T3 cells were used as the parental cell line and negative control, pCMV3-HER2 WT/S310F-mutant and pMH3-HER3 constructs were cotransfected into NIH3T3 cells. (B) Different ability of pertuzumab to inhibit HER2 phosphorylation signalling in WT/S310F-mutant HER2 cell lines after stimulation with or without HRG. NIH3T3 cells co-expressing WT/S310F HER2 and HER3 were incubated with culture medium or pertuzumab/trastuzumab for 2 h and then stimulated with or without HRG. Western blotting was performed using antibodies for the indicated proteins. (C) HER2 expression levels of MCF7 cells and MCF7 cells transfected with WT (MCF7-HER2 WT) or S310F-mutant (MCF7-HER2 S310F) HER2 gene, respectively. (D) Cell viability of MCF7, MCF7-HER2 WT and MCF7-HER2 S310F cell lines treated with culture medium (basal), HRG, HRG+pertuzumab or HRG+trastuzumab. Cell viability (%) is represented as the mean ± SD. One-way analysis of variance (ANOVA) was applied for statistical comparisons on multiple groups, ***p < 0.001.
Abbreviation: NS, no significance.