Figures & data
Figure 1 Clinical significance of CUL4B expression in GC patients. CUL4B mRNA expression based on Oncomine datasets: DErrico gastric (A) and Chen gastric (B) no value (0), diffuse gastric adenocarcinoma (1), gastric adenocarcinoma (2), gastric intestinal type adenocarcinoma (3) and gastric mixed adenocarcinoma (4). (C) Real-time PCR analysis of CUL4B mRNA expression in 50 human GC tissues and corresponding normal mucosa. **P<0.01. (D) Western blot analysis of CUL4B protein expression in 4 representative paired GC tissue samples. Immunohistochemical staining for CUL4B in adjacent normal mucosa (E) and GC tissues (F). Original magnification: 200×.
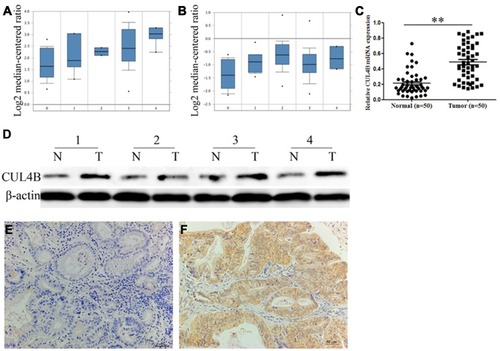
Table 1 CUL4B Expression in Normal Mucosa and GC Tissues
Table 2 Association Between CUL4B Expression and Clinicopathological Features
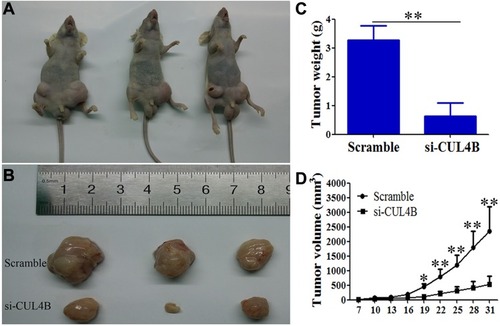
Figure 2 Comparison of disease-free survival (P=0.013) and overall survival (P=0.017) in patients with CUL4B positive and CUL4B negative tumors.
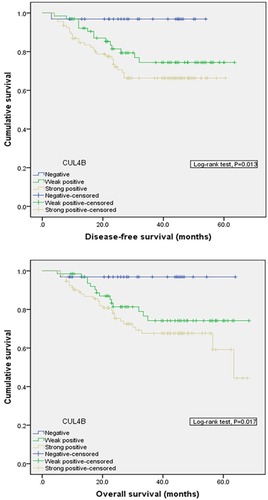
Table 3 Univariate and Multivariate Cox Proportional Hazard Models for Disease-Free Survival
Table 4 Univariate and Multivariate Cox Proportional Hazard Models for Overall Survival
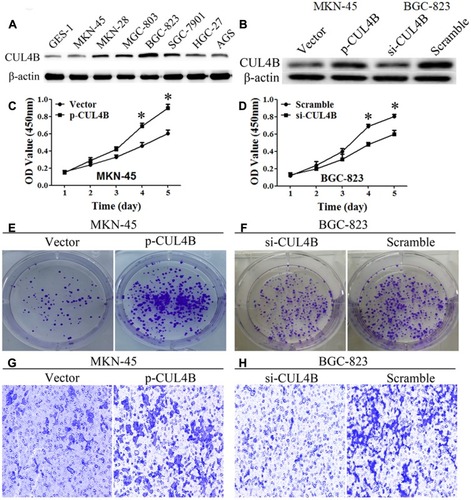
Figure 3 CUL4B regulates GC cell behaviors in vitro. CUL4B protein levels in GC cell lines (A) and in CUL4B overexpression and knockdown MKN-45 and BGC-823 cell lines, respectively (B). Overexpression or knockdown of CUL4B elevated or inhibited GC cells proliferation (C, D), colony formation (E, F), and invasion ability (G, H), compared with their control groups, respectively (*P<0.05). (G, H) Original magnification: 200×.