Figures & data
Figure 1 EMX2OS was lowly expressed in cells and tissues of PCa. (A) EMX2OS expression in PCa tissues and paracancerous tissues was detected by qPCR. (B) EMX2OS expression in PCa cell lines (LNCap, DU145 and PC3) and normal prostate epithelial cells (RWPE-1) was detected by qPCR. **p<0.01.
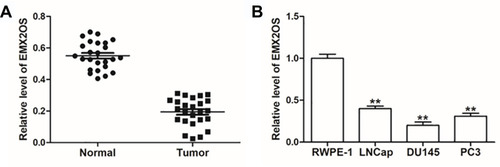
Figure 2 EMX2OS inhibited proliferation, migration and invasion of DU145 cells and activated cGMP-PKG pathway. The DU145 cells were transfected with EMX2OS overexpression vector, EMX2OS siRNA and respective negative controls, following transfection for 48 h, (A) The EMX2OS overexpression and inhibition efficiencies were detected by qPCR, and cell proliferation (B), invasion (C) and migration (D) were detected with CCK-8 and Transwell assay. The protein levels of PKG1 and PKG2 (E), and the cGMP concentration (F) in supernatant were detected by Western blotting and ELISA. *p<0.05, **p<0.01.
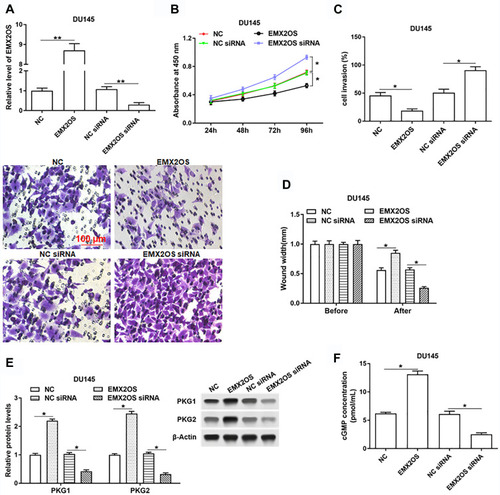
Figure 3 TCF12 directly bound to EMX2OS promoter. ChIPBase was used to predict the potential binding sites between TCF12 and EMX2OS. (A) Graphical representations of TCF12 binding regions’ detailed information. TCF12 binding regions were presented in wathet and the exons of EMX2OS were presented in prasinous. The transcription direction was indicated by wathet arrow. The binding motif was indicated in red bases. (B) Relative luciferase activities were detected by luciferase reporter gene assay. These reporters consisted of different inserts cloned from EMX2OS (indicated in blue), which were located upstream of the luciferase reporters and downstream of TCF12 sequence. (C) Anti-TCF12 was used to perform ChIP experiments in DU145 cells, and the input chromatin template (20%) was used as a positive control and IgG-precipitated template was used as a specificity control, and the enrichment of EMX2OS promoter sequence was detected by qPCR. **p<0.01.
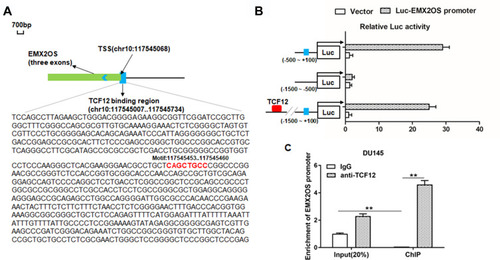
Figure 4 TCF12 was lowly expressed in cells and tissues of PCa and regulated cell proliferation, invasion and migration of DU145 cells. (A) TCF12 expression in PCa tissues and paracancerous tissues was detected by Western blotting. (B) TCF12 expression in PCa cell lines (LNCap, DU145 and PC3) and normal prostate epithelial cells (RWPE-1) was detected by Western blotting. The DU145 cells were transfected with TCF12 overexpression vector, TCF12 siRNA and respective negative controls, following transfection for 48 h, (C) the TCF12 overexpression and inhibition efficiencies were detected by Western blotting, and cell proliferation (D), invasion (E) and migration (F) were detected with CCK-8 and Transwell assay. *p<0.05, **p<0.01.
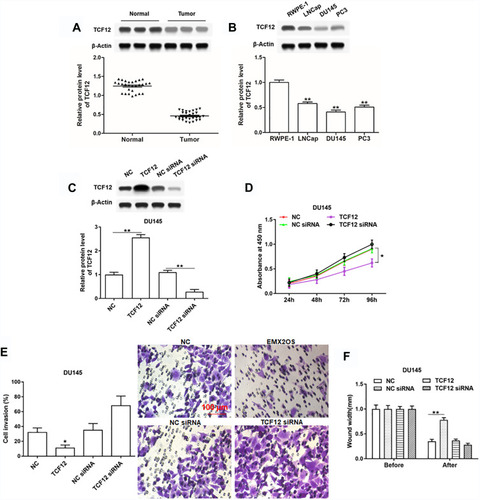
Figure 5 EMX2OS directly bound with FUS protein in DU145 cells. (A) The FUS binding motif was shown. (B) Schema of FUS binding sites in EMX2OS sequence. Red fonts indicated the binding motifs; blue and yellow fonts indicated the mutated nucleotides in the binding motifs. (C) Binding of EMX2OS and FUS was validated by FUS-antibody-based RNA-IP assay. (D) Binding of EMX2OS and FUS was validated by EMX2OS-probe-based RNA pull-down assay. (E) Binding of EMX2OS and FUS was validated by RNA pull-down assay using WT or MUT biotinylated EMX2OS transcripts. *p<0.05, **p<0.01, #p<0.05 versus MUT-both group.
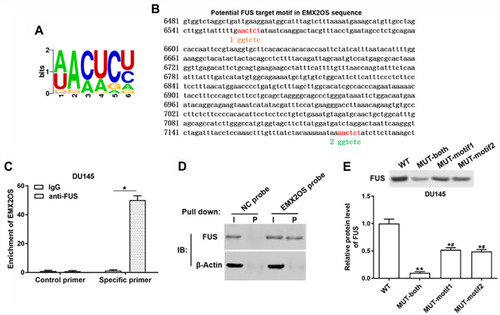
Figure 6 EMX2OS played synergy effect with FUS protein in DU145 cells. The DU145 cells were transfected with FUS overexpression vector alone or together with EMX2OS overexpression vector, or transfected with FUS siRNA alone or together with EMX2OS siRNA, following transfection for 48 h, (A) the FUS overexpression and inhibition efficiencies were detected by Western blotting, and the protein levels of PKG1 and PKG2 (B), and the cGMP concentration (C) in supernatant were detected by Western blotting and ELISA. Cell proliferation (D), migration (E) and invasion (F) were detected with CCK-8 and Transwell assay. *p<0.05, **p<0.01.
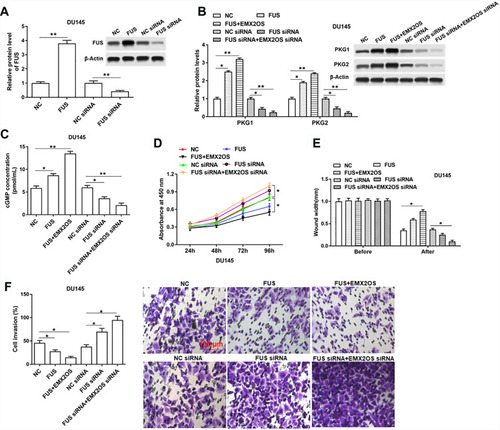
Figure 7 The effect of FUS and EMX2OS on DU145 cell behaviors was likely to be achieved via the cGMP-PKG pathway. The DU145 cells were transfected with EMX2OS and FUS overexpression vectors, following transfection for 48 h, (D)-DT-2 (2 μM) which was an inhibitor of cGMP-PKG pathway was used to treat cells for 6 h, and then the protein levels of PKG1 and PKG2 (A), and the cGMP concentration (B) in supernatant were detected by Western blotting and ELISA. Cell migration (C), invasion (D) and proliferation (E) were detected with Transwell and CCK-8 assay. *p<0.05, **p<0.01.
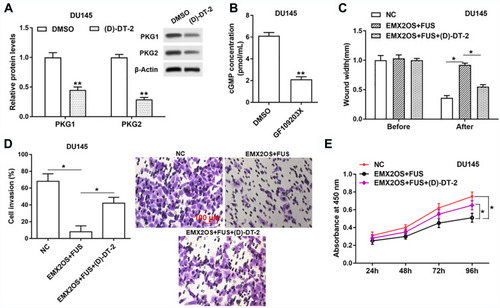
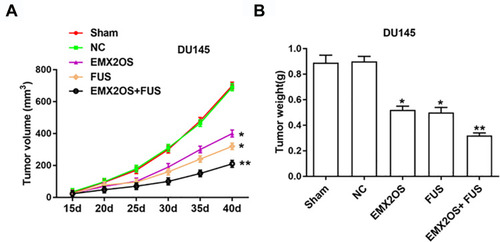
Figure 8 EMX2OS and FUS synergistically inhibited tumor growth in vivo. We generated xenograft mouse models by subcutaneous injection of DU145 cells (2 × 106) treated with EMX2OS or/and FUS overexpression vectors into nude mice, and tumor volumes (A) and weights (B) were measured every five days. *p<0.05, **p<0.01.