Figures & data
Figure 1 The identification of circNELL2. (A) circNELL2 was back-spliced by exon 14 and exon 15 of the neural EGFL like 2 (NELL2) gene with the length of 294 nucleotides. The existence of circNELL2 were confirmed by sanger sequencing, the arrow shows the head-to-tail splicing junction site of circNELL2. (B) After RNase R treatment, the expression of linear RPL13 mRNA and circNELL2 were detected using qPCR. (C) qRT-PCR analysis of RPL13 mRNA and circNELL2 in cardiomyocytes after treatment with Actinomycin D for 1, 2, 3, 4, 5 h, respectively. (D) The existence of circNELL2 was validated by RT-PCR. Divergent primers amplified circNELL2 in cDNA but not genomic DNA (gDNA). (E) qPCR was performed to evaluate the expression of circNELL2 in ESCC tissues. *P< 0.05, **P< 0.01.
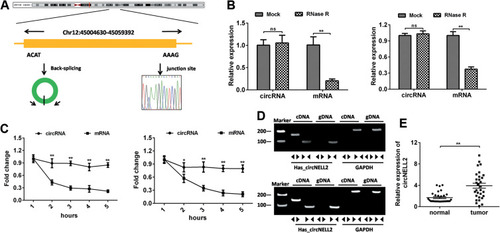
Figure 2 circNELL2 knock down attenuates cell proliferation, colony formation, migration and invasion of ESCC cell lines. (A) qPCR was performed to evaluate the expression level of circNELL2 in ESCC cell under different treatment. Cell viability of OE33 and FLO-1 cells was investigated using (B) EDU staining and (C) WST assay. (D) Clonability of OE33 and FLO-1 cells was investigated by colony-formation assay. (E) Migration ability of OE33 and FLO-1 cells were detected by transwell experiment. *P < 0.05.
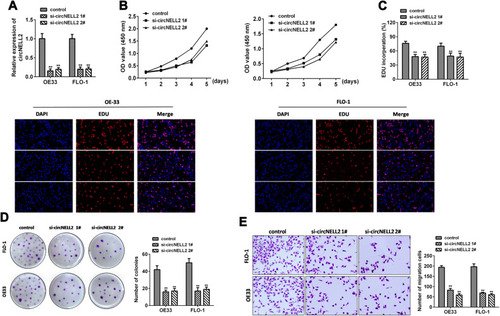
Figure 3 circNELL2 directly targets miR-127-5p. (A) circNELL2 was predicated as a potential target gene of miR-127-5p, wild and mutant type of targeting seed sequences between circNELL2 and miR-127-5p are shown. (B)Luciferase activities were used to investigate the targeting of circNELL2 and miR-127-5p. (C) qPCR was performed to evaluate the expression of miR-127-5p in response to circNELL2 overexpression or knock down. (D) FISH experiment using the probe of miR-127-5p and circNELL2 were carried out to evaluate the co-location of them. (E) RNA pull down using the specific probe of miR-127-5p was performed to evaluate the interaction between miR-485-3p and circNELL2. (F) Pearson analysis was performed to evaluate the correlation between miR-127-5p and circNELL2. *P < 0.05.
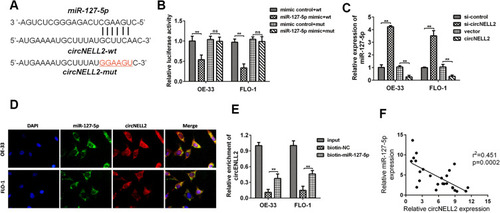
Figure 4 Restoration of miR-127-5p and knock down of RELA significantly rescues circNELL2-mediated regulation of cellular effects. (A) qPCR was performed to evaluate the expression of RELA under different treatment. OE33 and FLO-1 cell viability was detected using (B) WST assay and (C, D) EDU staining assay. (E, F) Clonability of OE33 and FLO-1 cells was investigated by colony-formation assay. (G, H) Migration ability of OE33 and FLO-1 cells were detected by transwell experiment. *P < 0.05.
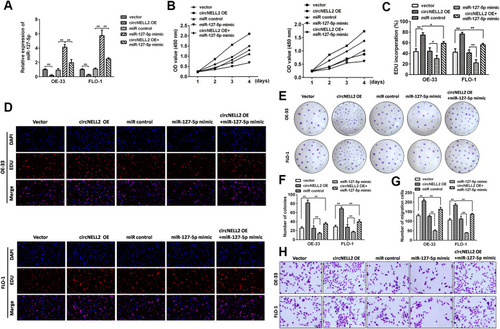
Figure 5 miR-127-5p directly targets CDC6 in ESCC cells. (A) The potential target gens were predicted by Targetscan, MiRDB. (B) CDC6 was predicated as a potential target gene of miR-127-5p, wild and mutant type of targeting seed sequences between CDC6 and miR-127-5p are shown. (C) Luciferase activities were used to investigate the targeting of CDC6 and miR-127-5p. (D, E) qPCR and Western blot assays were performed to evaluate the mRNA and protein expression of CDC6 in response to miR-127-5p overexpression or knock down. (F) RNA pull down was carried out to detect the binding of miR-127-5p and CDC6. (G) qPCR was used to assess the expression of CDC6. (H, I) Pearson analysis was performed to evaluate the correlation between miR-127-5p and CDC6 as well as CDC6 and circNELL2. *P < 0.05 vs miR control, mimic control+wt and biotin-nc.
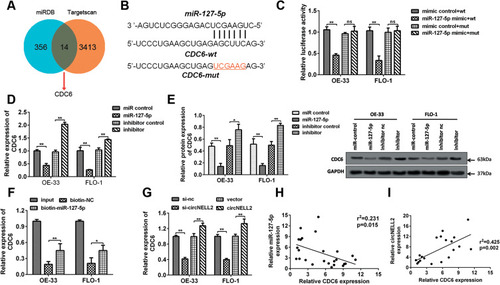
Figure 6 CDC6 is up-regulated in ESCC. qRT-PCR analysis of CDC6 in (A) ESCC tissues and (B) cell lines were assessed. (C) Analysis based on the data from starbase dataset was performed to show that CDC6 was up-regulated in ESCC tissues. (D) IHC was further carried out to confirmed the expression of CDC6 in tumor tissues. *P< 0.05, **P< 0.01.
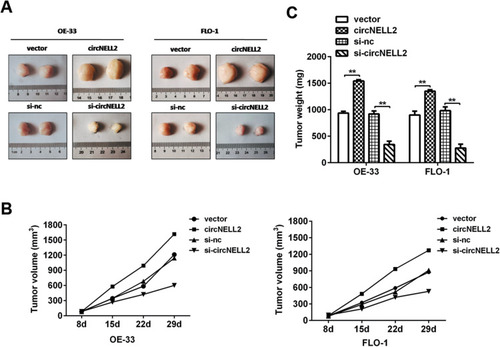
Figure 7 circNELL2 promote xenograft tumour growth in vivo. A xenograft tumour model was established by subcutaneously injecting OE33 and FLO-1 cells stably over-expressing or knocking down circNELL2 along with their blank control into the dorsal flank of nude mice. (A) Tumour image was shown in each group. (B) Tumour weights was weighed. (C) The growth curves of the tumors in each group was made. *P < 0.05.