Figures & data
Figure 1 The expression of circ_0000512 was enhanced in CRC tissues and cells, and correlated with poor clinical outcomes. (A) GSE126094 (10 pairs of CRC tissues and normal tissues) from the GEO database was analyzed to determine the expression of circ_0000512 in CRC tissues and normal tissues. (B) The expression of circ_0000512 was detected by qRT-PCR in CRC tissues and normal tissues. (C) The overall survival rate was measured between low and high circ_0000512 expression groups in CRC patients by Kaplan-Meier survival analysis. (D) The level of circ_0000512 was analyzed by qRT-PCR in CRC cells (SW480, HCT116, SW620, LoVo) and FHC cells. (E) The relative levels circ_0000512 and GAPDH were determined after treatment of RNase R by qRT-PCR. *P<0.05, ***P<0.001.
Abbreviations: CRC, colorectal cancer; GEO, Gene Expression Omnibus; qRT-PCR, quantitative real-time polymerase chain reaction; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
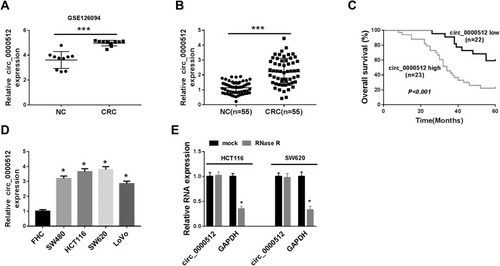
Figure 2 Downregulation of circ_0000512 repressed proliferation and facilitated apoptosis in CRC cells. (A) The abundance of circ_0000512 was examined by qRT-PCR in HCT116 and SW620 cells transfected with sh-NC, sh-circ#1, sh-circ#2, sh-circ#3. (B–G) HCT116 and SW620 cells were transfected with sh-NC or sh-circ#2. (B) Cell viability was assessed by CCK-8 analysis. (C) Colony formation assay was used to examine the number of colonies. (D) Flow cytometry was applied to determine the cell cycle distribution. (E and F) Flow cytometry analysis was utilized to measure the apoptosis rate. (G) The protein levels of Cyclin D1 and Cleaved Caspase-3 were examined by Western blot assay. *P<0.05, **P<0.01.
Abbreviations: CRC, colorectal cancer; qRT-PCR, quantitative real-time polymerase chain reaction; CCK-8, Cell Counting Kit-8.
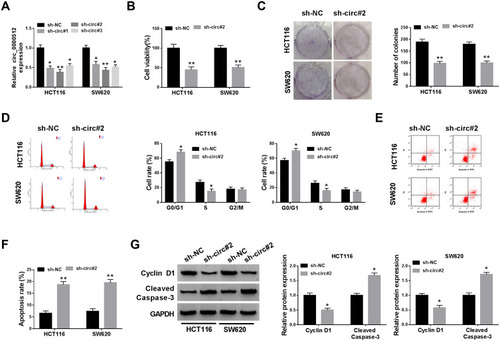
Figure 3 Circ_0000512 directly interacted with miR-296-5p in CRC cells. (A) The qRT-PCR assay determined the subcellular location of circ_0000512 in HCT116 and SW620 cells. (B) The potential target miRNAs of circ_0000512 were predicted by starBase and circinteractom. (C) The expression levels of miR-296-5p, miR-326 and miR-330-5p were detected by qRT-PCR in HCT116 and SW620 cells transfected with sh-NC or sh-circ#2. (D) The abundance of miR-296-5p was measured by qRT-PCR in HCT116 and SW620 cells transfected with miR-NC or miR-296-5p. (E) The putative binding sites between circ_0000512 and miR-296-5p were predicted by starBase. (F and G) The luciferase activity was measured in HCT116 and SW620 cells co-transfected with circ_0000512-WT or circ_0000512-MUT and miR-296-5p or miR-NC. (H and I) The enrichment of miR-296-5p and circ_0000512 was detected in HCT116 and SW620 cells incubated with anti-ago2 or anti-IgG by RIP assay. (J) The level of circ_0000512 was examined in HCT116 and SW620 cells transfected with Bio-miR-296-5p or Bio-NC by RNA pull-down assay. **P<0.01, ***P<0.001.
Abbreviations: miR-296-5p, microRNA-296-5p; CRC, colorectal cancer; qRT-PCR, quantitative real-time polymerase chain reaction; RIP, RNA immunoprecipitation.
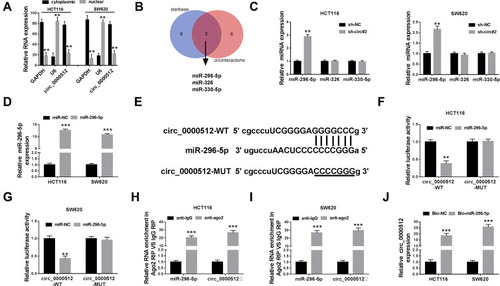
Figure 4 RUNX1 was a direct target of miR-296-5p in CRC cells. (A) The potential binding sites of RUNX1 and miR-296-5p were predicted by starBase. (B) The luciferase activity was tested in HCT116 and SW620 cells co-transfected with RUNX1 3ʹUTR-WT or RUNX1 3ʹUTR-MUT and miR-296-5p or miR-NC. (C and D) QRT-PCR and Western blot analyses were conducted to determine the mRNA and protein expression of RUNX1 in HCT116 and SW620 cells transfected with miR-296-5p or miR-NC. (E) The abundance of miR-296-5p was measured by qRT-PCR in HCT116 and SW620 cells transfected with anti-NC or anti-miR-296-5p. (F and G) The mRNA and protein levels of RUNX1 were measured in HCT116 and SW620 cells transfected with sh-NC, sh-circ#2, sh-circ#2 + anti-NC, or sh-circ#2 + anti-miR-296-5p. *P<0.05, **P<0.01.
Abbreviations: miR-296-5p, microRNA-296-5p; CRC, colorectal cancer; RUNX1, runt-related transcription factor 1; qRT-PCR, quantitative real-time polymerase chain reaction; 3ʹUTR, 3ʹuntranslated region.
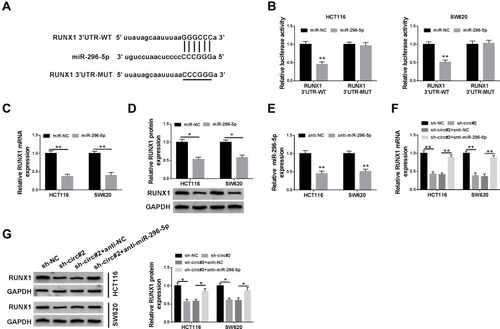
Figure 5 Knockdown of circ_0000512 suppressed proliferation and promoted apoptosis in CRC cells by upregulating miR-296-5p and downregulating RUNX1. (A) Western blot assay was performed to measure the protein expression of RUNX1 in HCT116 and SW620 cells transfected with vector or RUNX1. (B–F) HCT116 and SW620 cells were transfected with sh-NC, sh-circ#2, sh-circ#2 + anti-NC, sh-circ#2 + anti-miR-296-5p, sh-circ#2 + vector, sh-circ#2 + RUNX1. (B) CCK-8 assay was used to evaluate cell viability. (C) The number of colonies was determined by colony formation assay. (D) Cell cycle distribution was analyzed using flow cytometry. (E) Cell apoptosis was examined by flow cytometry. (F) The protein levels of Cyclin D1 and Cleaved Caspase-3 were measured by Western blot analysis. *P<0.05, **P<0.01.
Abbreviations: CRC, colorectal cancer; miR-296-5p, microRNA-296-5p; RUNX1, runt-related transcription factor 1; CCK-8, Cell Counting Kit-8.
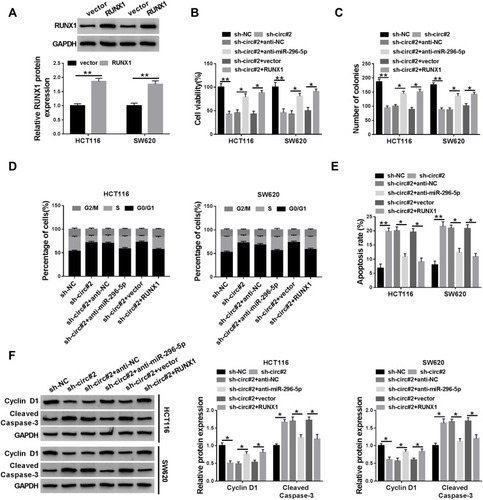
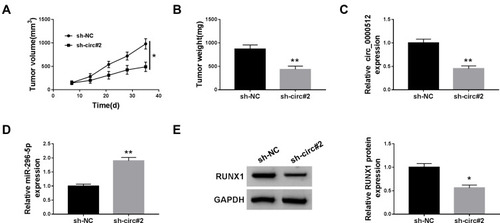
Figure 6 Deficiency of circ_0000512 inhibited tumor growth by enhancing miR-296-5p and decreasing RUNX1. Sh-NC or sh-circ#2-transfected SW620 cells were introduced into nude mice to establish mice xenograft model. (A and B) Tumor volume and weight were measured. (C and D) The expression of circ_0000512 and miR-296-5p was analyzed by qRT-PCR in tumor tissues. (E) Western blot assay was used to determine the protein abundance of RUNX1 in tumor tissues. *P<0.05, **P<0.01.
Abbreviations: miR-296-5p, microRNA-296-5p; RUNX1, runt-related transcription factor 1; qRT-PCR, quantitative real-time polymerase chain reaction.