Figures & data
Table 1 Sequences Used for qRT-PCR
Figure 1 CTBP1-AS2 was highly expressed in GC and was associated with poor prognosis of patients. (A) CTBP1-AS2 expression in GC samples was analyzed using GEPIA database. (B) qRT-PCR was performed for detecting CTBP1-AS2 expression in GC tissues from 37 patients. (C–E) The relationships between CTBP1-AS2 expression level and patient’s TNM stage (C), tumor size (D) and degree of differentiation (E). (F) Kaplan–Meier method was employed for analyzing the relationship between CTBP1-AS2 expression and post-progression survival time of GC patients. *P < 0.05, **P < 0.01, and ***P < 0.001.
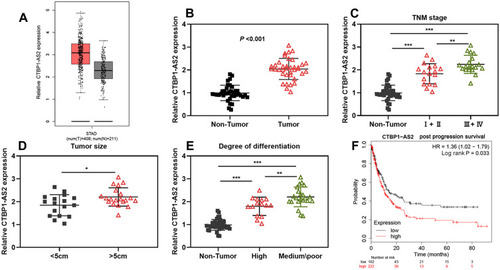
Figure 2 CTBP1-AS2 facilitated GC cell proliferation and inhibited apoptosis. (A) qRT-PCR was utilized for detecting CTBP1-AS2 expression level in human gastric mucosal cell line (GES-1) and GC cell lines (SNU-1, NCI-N87, HGC-27 and AGS). (B) Transfection efficiency of CTBP1-AS2 overexpression plasmids and si-RNA#/si-RNA#2 targeting CTBP1-AS2 was detected by qRT-PCR. (C–D) CCK-8 assay (C) and EdU assay (D) were used to detect GC cell proliferation. (E) Flow cytometry analysis was used to detect apoptosis of GC cells. (F) Western blot was conducted to detect the effect of CTBP1-AS2 on Bcl-2 and Bax expressions in GC cells after transfection. *P < 0.05, **P < 0.01, and ***P < 0.001.
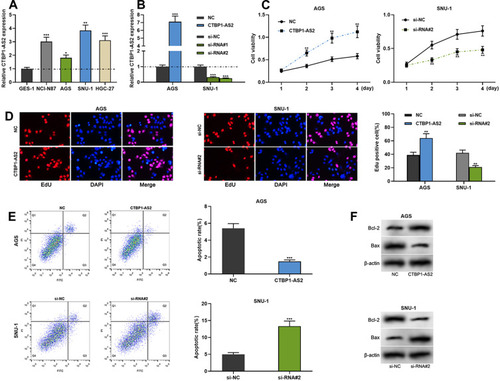
Figure 3 CTBP1-AS2 targetedly regulated miR-139-3p. (A) Bioinformatics analysis predicted the binding site between CTBP1-AS2 and miR-139-3p. (B) Dual-luciferase reporter experiment was conducted to detect the binding relationship between CTBP1-AS2 and miR-139-3p. (C–D) RNA pull-down assay and Ago2-RIP assay were used to verify the relationship between CTBP1-AS2 and miR-139-3p in GC cells. (E) After overexpression or knockdown of CTBP1-AS2, qRT-PCR was utilized for detecting miR139-3p expression in GC cells. (F–G) qRT-PCR was used for detecting miR-139-3p expression in GC tissues (F), and the correlation between the miR-139-3p and CTBP1-AS2 was calculated (G). ***P < 0.001.
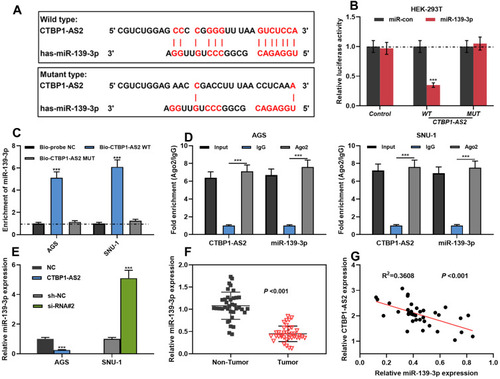
Figure 4 CTBP1-AS2 promoted cell proliferation and inhibited apoptosis by adsorbing miR-139-3p. AGS cells were transfected with CTBP1-AS2 overexpression plasmids or CTBP1-AS2 overexpression plasmids + miR-139-3p mimics; SNU-1 cells were transfected with CTBP1-AS2 siRNA or CTBP1-AS2 siRNA + miR-139-3p inhibitors. (A) qRT-PCR was used for detecting miR-139-3p expression in GES-1 cells and GC cell lines. (B) qRT-PCR was utilized for detecting the expression of miR-139-3p after the co-transfection. (C–E) CCK-8 assay (C), EdU assay (D) and flow cytometry analysis (E) were performed for detecting the proliferation and apoptosis of SNU-1 and AGS cells after co-transfection. (F) Western blot was used for detecting the regulatory effects of CTBP1-AS2 and miR-139-3p on Bcl-2 and Bax expression. *P < 0.05, **P < 0.01, and ***P < 0.001.
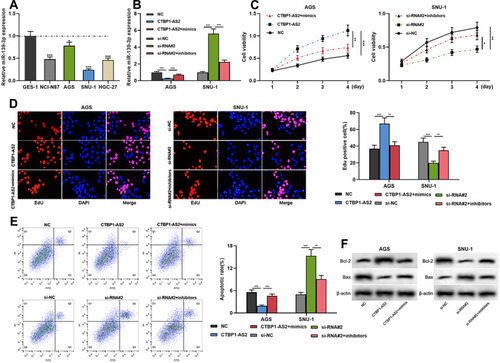
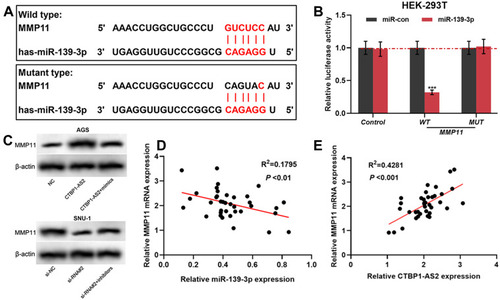
Figure 5 MMP11 was a target gene of miR-139-3p, and could be positive regulated by CTBP1-AS2. (A) TargetScan predicted the binding sequence between MMP11 and miR-139-3p. (B) Dual-luciferase reporter experiment was conducted to detect the binding relationship between MMP11 and miR-139-3p. (C) Western blot was utilized for detecting the regulatory effects of miR-139-3p (C) and CTBP1-AS2 (D) on MMP11 protein expression level. (D–E) qRT-PCR was used to detect the correlation between MMP11 and miR-139-3p expression in GC samples. ***P < 0.001.