Figures & data
Table 1 Correlation Between PCED1B-AS1 Expression and Clinicopathologic Features of ccRCC Patients
Figure 1 PCED1B-AS1 was up-regulated in ccRCC tissues and cell lines and was associated with adverse prognosis of the patients. (A) In the GEPIA database, PCED1B-AS1 was highly expressed in ccRCC tissues (red: cancerous tissues; gray: normal tissues). (B) GEPIA database was used to analyze the relationship between the expression of PCED1B-AS1 in ccRCC patients and the overall survival time of patients. (C) qRT-PCR was employed to detect the expression level of PCED1B-AS1 in ccRCC tissue and normal kidney tissue. (D) qRT-PCR was used to detect the expression of PCED1BAS1 in normal renal tubular epithelial cell line HK-2 and ccRCC cell lines (786-O, A498, ACHN and Caki-1). *P<0.05, **P <0.01, and ***P < 0.001.
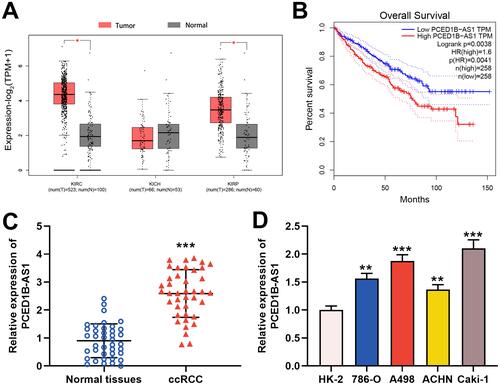
Figure 2 PCED1B-AS1 promoted the proliferation and migration of ccRCC cells. (A) qRT-PCR was used to detect the transfection efficiency of si-PCED1B-AS1. (B, C) The effect of silencing PCED1B-AS1 on the proliferation of ccRCC cells was determined by CCK-8 and EdU experiments. (D) Wound healing experiment was employed to detect the effect of silencing PCED1B-AS1 on the migration of ccRCC cells. (E) Western blotting was used to detect the expression levels of EMT-related proteins. *P < 0.05; **P < 0.01; ***P < 0.001.
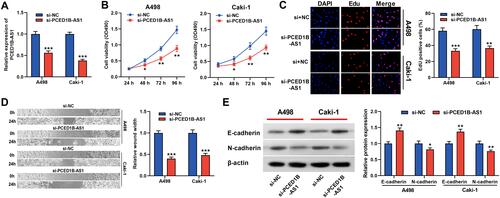
Figure 3 PCED1B-AS1 acted as a ceRNA of miR-484 in ccRCC. (A) qRT-PCR was used to detect the subcellular location of PCED1B-AS1 in A498 and Caki-1 cells. (B) Bioinformatics was used to predict the binding site for miR-484 on PCED1B-AS1 sequence. (C) MiR-484 mimics or NC mimics and PCED1B-AS1-WT luciferase reporter or PCED1B-AS1-MUT luciferase reporter were co-transfected into A498 and Caki-1 cells, and the relative luciferases activity in each group was detected. (D) The direct interaction between PCED1B-AS1 and miR-484 was detected by RIP method. (E) qRT-PCR was used to detect the relative expression level of miR-484 in ccRCC tissue and normal kidney tissue. (F) Pearson’s correlation analysis was used to evaluate the correlation between PCED1B-AS1 and miR-484 expression levels in ccRCC tissues. (G) qRT-PCR was used to detect the expression of miR-484 in normal renal tubular epithelial cell line HK-2 and ccRCC cell lines (786-O, A498, ACHN and Caki-1). **P < 0.01, and ***P < 0.001.
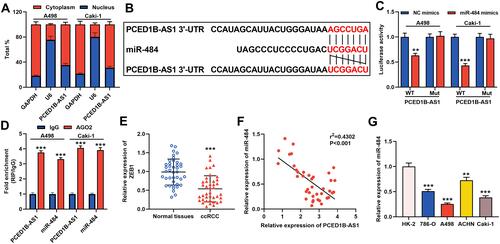
Figure 4 miR-484 played a tumor-suppressive role in the progress of ccRCC. (A) qRT-PCR was used to detect miR-484 expression level of A498 and Caki-1 cells after miR-484 mimics or NC mimics were transfected. (B, C) CCK-8 and EdU assays were used to detect the effect of miR-484 on the proliferation of A498 and Caki-1 cells. (D) The effect of miR-484 on the migration of A498 and Caki-1 cells was detected by the wound healing experiment. (E) Western blotting was used to detect the expression level of EMT-related proteins. *P < 0.05, **P < 0.01, and ***P < 0.001.
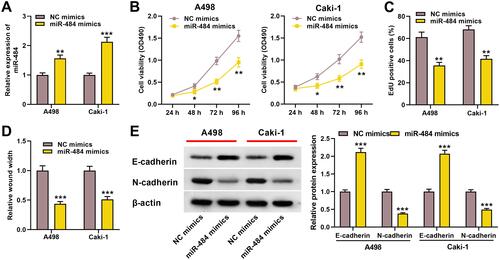
Figure 5 ZEB1 was a direct target gene of miR-484. (A) Bioinformatics analysis was used to predict the binding site of miR-484 on the ZEB1 3ʹ UTR sequence. (B) MiR-484 mimics or NC mimics and ZEB1-WT luciferase reporter or ZEB1-MUT luciferase reporter were co-transfected into A498 and Caki-1 cells, and the relative luciferase activity of the cells in each group was measured. (C) Western blotting was used to detect ZEB1 protein expression in miR-484 mimics-transfected cells. (D) qRT-PCR was used to detect the relative expression level of ZEB1 in ccRCC tissue and normal kidney tissue. (E) Pearson’s correlation analysis was used to evaluate the correlation between PCED1B-AS1 and ZEB1 expression levels in ccRCC tissues. **P < 0.01, and ***P < 0.001.
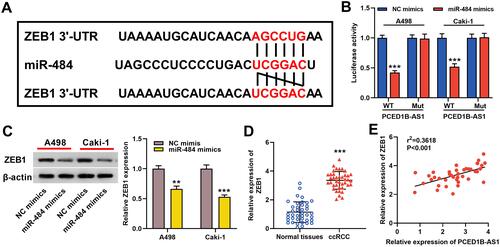
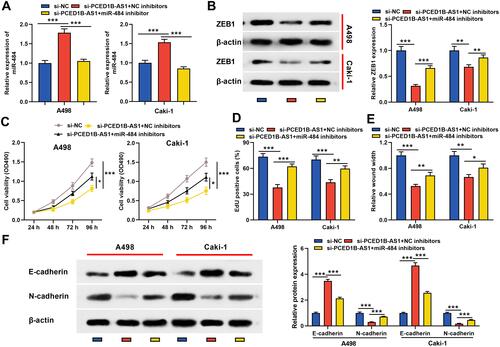
Figure 6 PCED1B-AS1 played a carcinogenic role in ccRCC cells through the miR-484/ZEB1 axis. miR-484 inhibitors or NC inhibitors were co-transfected into A498 and Caki-1 cells with PCED1B-AS1 silenced. (A) qRT-PCR was employed to detect miR-484 expression. (B) Western blotting was used to detect ZEB1 protein expression. (C, D) CCK-8 and EdU were used to detect the proliferation of A498 and Caki-1 cells. (E) Wound healing test was used to detect the migration of A498 and Caki-1 cells. (F) Western blotting was used to detect the expression level of EMT-related proteins. *P < 0.05, **P < 0.01, and ***P < 0.001.