Figures & data
Figure 1 Circ_0067835 was upregulated in CRC. (A) The expression of circ_0067835 was detected by qRT-PCR in CRC tissues (n=39) and adjacent normal tissues (ANT) (n=39). (B) ROC curve and AUC values were analyzed for circ_0067835 in CRC patients. (C) The expression of circ_0067835 was detected by qRT-PCR in the serum of CRC patients with (n=19) or without (n=19) radiation treatment. (D) The expression of circ_0067835 in exosome was detected by qRT-PCR in the serum of CRC patients (n=39) and normal patients (n=31). (E) The expression of circ_0067835 in exosome was detected by qRT-PCR in the serum of CRC patients with (n=19) or without (n=19) radiation treatment. (F) The size of cellular exosome diameters was detected by NTA. **P<0.01, ***P<0.001.
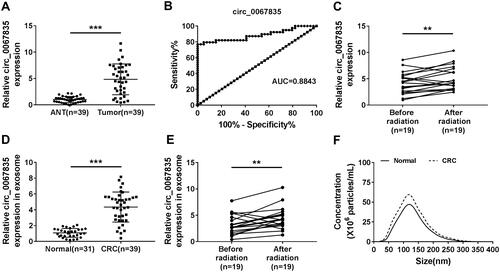
Figure 2 Exosome-mediated circ_0067835 knockdown inhibited cell proliferation, cell cycle progression, and enhanced apoptosis and radiosensitivity in vitro. (A) Circ_0067835 expression in SW620 and HCT116 cells and the normal colon epithelial cells (FHC) was detected by qRT-PCR. (B) qRT-PCR analyzed circ_0067835 expression in SW620 and HCT116 cells transfected with sh-circ_0067835 or sh-NC. (C and D) The viability of SW620 and HCT116 cells treated with sh-circ_0067835-exo or negative control was detected by CCK-8 assay. (E and F) The survival fraction of SW620 and HCT116 cells treated with 0 Gy, 1 Gy, 2 Gy, 4 Gy, 8 Gy IR and sh-circ_0067835-exo or negative control was detected by colony formation assay. (G) Cell cycle distribution was measured by flow cytometry assay in SW620 and HCT116 cells treated with or without 4 Gy IR and sh-circ_0067835-exo or negative control. (H) The caspase-3 activity was tested by caspase-3 activity assay in SW620 and HCT116 cells treated with or without 4 Gy IR and sh-circ_0067835-exo or negative control. (I) Flow cytometry assay was used to detect the apoptosis rate in SW620 and HCT116 cells treated with or without 4 Gy IR and sh-circ_0067835-exo or negative control. *P<0.05, **P<0.01, ***P<0.001.
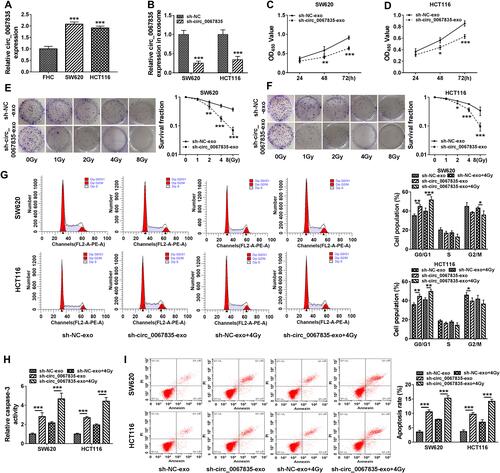
Figure 3 Circ_0067835 targeted miR-296-5p and miR-296-5p overexpression suppressed cell proliferation, cell cycle progression, and promoted apoptosis and radiosensitivity. (A) The miRNA containing the binding sites of circ_0067835 was predicted by online database Circinteractome. (B) Dual-luciferase reporter assays analyzed the relationship between miR-296-5p and circ_0067835. (C) Relative expression level of miR-296-5p in CRC tissues and adjacent normal tissues (ANT) was detected by qRT-PCR. (D) MiR-296-5p expression in the serum of CRC patients with (n=19) or without (n=19) radiation treatment was detected by qRT-PCR. (E) Pearson Correlation Coefficient analyzed the relationship between the expression of miR-296-5p and circ_0067835. (F) MiR-296-5p expression was detected by qRT-PCR in SW620 and HCT116 cells transfected with sh-circ_0067835 or sh-NC. (G) CCK-8 assay was used to detect the viability of SW620 and HCT116 cells transfected with miR-296-5p mimics or miR-NC mimics. (H) Colony formation assay was applied to determine the survival fraction of SW620 and HCT116 cells treated with 0 Gy, 1 Gy, 2 Gy, 4 Gy, 8 Gy IR and transfected with miR-296-5p or miR-NC. (I and K) Flow cytometry assay was used to assess cell cycle distribution in SW620 and HCT116 cells treated with 4 Gy IR and transfected with miR-296-5p or miR-NC. (J) The caspase-3 activity was detected by caspase-3 activity assay in SW620 and HCT116 cells treated with 4 Gy IR and transfected with miR-296-5p or miR-NC. *P<0.05, **P<0.01, ***P<0.001.
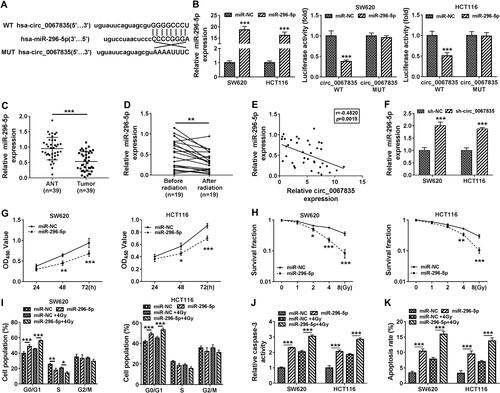
Figure 4 MiR-296-5p targeted IGF1R and circ_0067835 modulated IGF1R expression through miR-296-5p. (A) StarBase online tool was applied to predict the genes possessing the binding sequence of miR-296-5p. (B) Dual-luciferase reporter assays analyzed the relationship between IGF1R and miR-296-5p in SW620 and HCT116 cells. (C and D) Relative expression of IGF1R at both mRNA and protein levels in CRC tissues and adjacent normal tissues (ANT) were respectively detected by qRT-PCR and Western blot. (E and F) Relative mRNA and protein levels of IGF1R in SW620 and HCT116 cells and normal cells (FHC) were respectively quantified by qRT-PCR and Western blot. (G) IGF1R mRNA expression in the serum of CRC patients with (n=19) or without (n=19) radiation treatment was detected by qRT-PCR. (H) The relationship between the expression of miR-296-5p and IGF1R was analyzed by Pearson Correlation Coefficient in CRC tissues. (I) MiR-296-5p expression was determined by qRT-PCR in SW620 and HCT116 cells transfected with anti-miR-296-5p or anti-NC. (J and K) The expression of IGF1R at both mRNA and protein levels was respectively determined by qRT-PCR and Western blot in SW620 and HCT116 cells transfected with miR-296-5p mimics, anti-miR-296-5p or negative controls. (L and M) The IGF1R mRNA and protein expression were respectively detected by qRT-PCR and Western blot in SW620 and HCT116 cells transfected with sh-circ_0067835 or sh-NC, sh-circ_0067835+anti-miR-296-5p or sh-circ_0067835+anti-NC. **P<0.01, ***P<0.001.
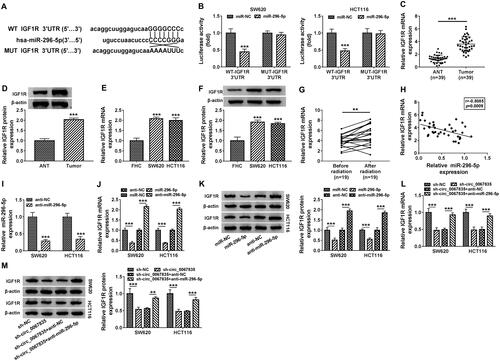
Figure 5 Knockdown of circ_0067835-regulated cell proliferation, cell cycle progression, apoptosis and radiosensitivity in vitro by up-regulating miR-296-5p. (A) MiR-296-5p expression was determined by qRT-PCR in SW620 and HCT116 cells transfected with sh-circ_0067835 or sh-NC, sh-circ_0067835+anti-miR-296-5p or sh-circ_0067835+anti-NC. (B) CCK-8 assay was employed to evaluate the viability of SW620 and HCT116 cells transfected with sh-circ_0067835 or sh-NC, sh-circ_0067835+anti-miR-296-5p or sh-circ_0067835+anti-NC. (C) Colony formation assay was used to assess the survival fraction of SW620 and HCT116 cells treated with 0 Gy, 1 Gy, 2 Gy, 4 Gy, 8 Gy IR and transfected with sh-circ_0067835 or sh-NC, sh-circ_0067835+anti-miR-296-5p or sh-circ_0067835+anti-NC. (D) Flow cytometry assay was used to detect the cell cycle distribution in SW620 and HCT116 cells transfected with sh-circ_0067835 or sh-NC, sh-circ_0067835+anti-miR-296-5p or sh-circ_0067835+anti-NC. (E) Caspase-3 activity assay was used to detect the caspase-3 activity in SW620 and HCT116 cells transfected with sh-circ_0067835 or sh-NC, sh-circ_0067835+anti-miR-296-5p or sh-circ_0067835+anti-NC. (F) Flow cytometry assay was used to detect cell apoptosis rate in SW620 and HCT116 cells transfected with sh-circ_0067835 or sh-NC, sh-circ_0067835+anti-miR-296-5p or sh-circ_0067835+anti-NC. **P<0.01, ***P<0.001.
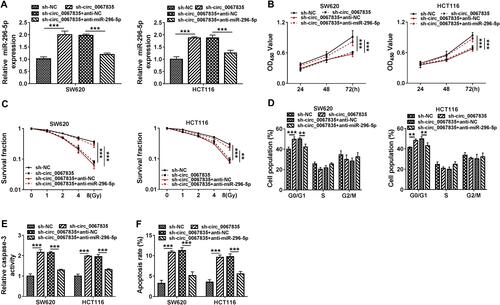
Figure 6 IGF1R was an important mediator of circ_0067835 function in CRC cells. (A and B) The expression of IGF1R mRNA and protein was tested by qRT-PCR and Western blot in SW620 and HCT116 cells transfected with IGF1R overexpression plasmid (IGF1R) or pcDNA. (C and D) The expression of IGF1R mRNA and protein was respectively detected by qRT-PCR and Western blot in SW620 and HCT116 cells transfected with sh-circ_0067835, sh-circ_0067835+IGF1R overexpression plasmid or negative controls. (E) CCK-8 assay was used to evaluate the viability of SW620 and HCT116 cells transfected with sh-circ_0067835, sh-circ_0067835+IGF1R or negative controls. (F) Colony formation assay was employed to assess the proliferation of SW620 and HCT116 cells treated with 0 Gy, 1 Gy, 2 Gy, 4 Gy, 8 Gy IR and transfected with sh-circ_0067835, sh-circ_0067835+IGF1R or negative controls. (G) Flow cytometry assay was used to detect the cell cycle distribution in SW620 and HCT116 cells transfected with sh-circ_0067835, sh-circ_0067835+IGF1R or negative controls. (H) Caspase-3 activity assay was used to detect the caspase-3 activity in SW620 and HCT116 cells transfected with sh-circ_0067835, sh-circ_0067835+IGF1R or negative controls. (I) Flow cytometry assay was used to detect cell apoptosis rate in SW620 and HCT116 cells transfected with sh-circ_0067835, sh-circ_0067835+IGF1R or negative controls. *P<0.05, **P<0.01, ***P<0.001.
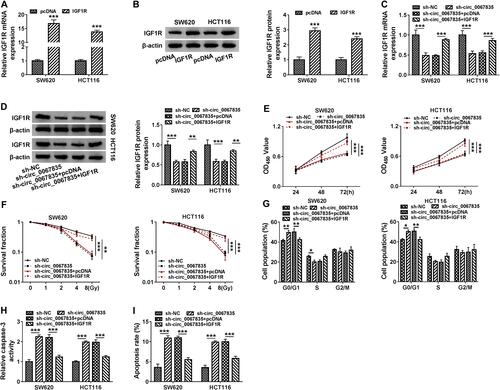
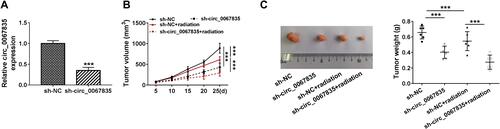
Figure 7 Circ_0067835 knockdown weakened tumor growth and enhanced cell radiosensitivity in vivo. (A) The expression of circ_0067835 was examined by qRT-PCR in sh-circ_0067835- or sh-NC-transduced SW620 tumors. (B) Tumor volumes were monitored every 5 days after injection of SW620 cells transfected with sh-circ_0067835 or sh-NC and treatment with or without 4 Gy IR. (C) The effects between circ_0067835 silencing and radiation treatment on tumor weight was measured. ***P<0.001.