Figures & data
Figure 1 Heatmap of differentially expressed genes (DEGs) between samples of human epithelial ovarian cancer cell line TOV112D untreated and treated with NSC319726.
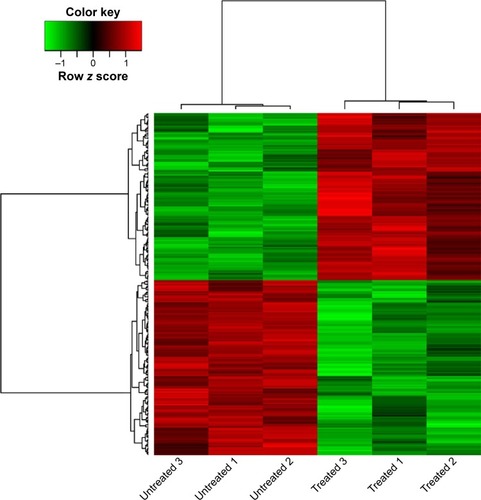
Table 1 KEGG pathways impacted by differentially expressed genes (DEGs) in samples of human epithelial ovarian cancer cell line TOV112D treated with NSC319726
Table 2 The results of functional annotation of differentially expressed genes
Figure 2 The constructed protein–protein interaction network of differentially expressed genes (DEGs).
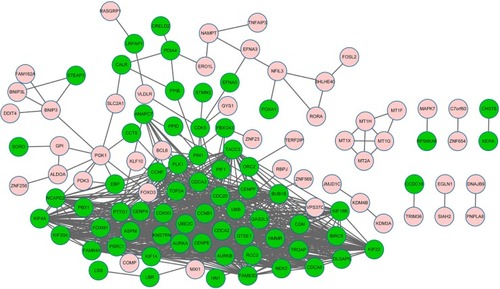
Figure 3 The constructed subnetwork of differentially expressed genes (DEGs).
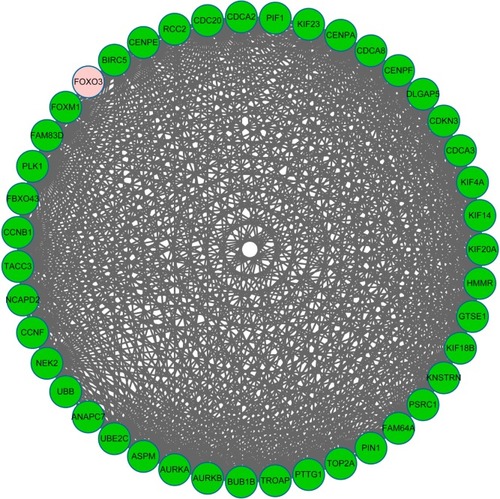
Table 3 Gene Ontology (GO) functional enrichment analysis of differentially expressed genes (DEGs) in the subnetwork (top 4)
