Figures & data
Figure 1 Binding and intracellular signaling of SLAMF7 receptors.
Abbreviations: EAT-2, Ewing’s sarcoma-associated transcript 2; ITSM, immunoreceptor tyrosine-based switch motif; NK, natural killer; SLAMF7, signaling lymphocytic activation molecule F7; TM, transmembrane domain; C2 and V, Ig superfamily domains.
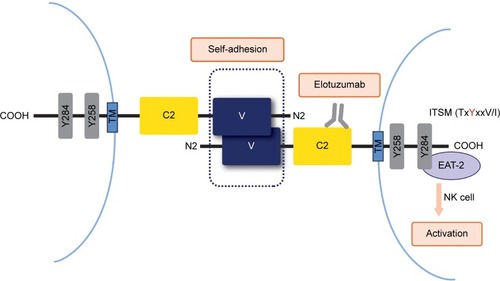
Figure 2 Dual mechanism of action of elotuzumab to induce myeloma cell death.
Abbreviations: ADCC, antibody-dependent cell-mediated cytotoxicity; EAT-2, Ewing’s sarcoma-associated transcript 2; NK, natural killer; SLAMF7, signaling lymphocytic activation molecule F7.
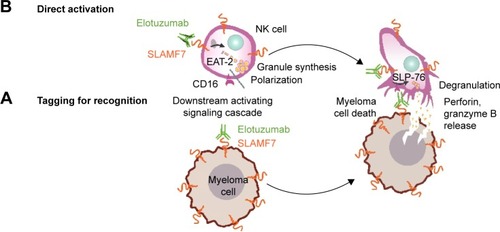
Figure 3 SLAMF7 saturation of BM CD38+ cells.
Abbreviations: BM, bone marrow; Emax, maximum effect; MM, multiple myeloma; RRMM, relapsed and/or refractory MM; SLAMF7, signaling lymphocytic activation molecule F7.
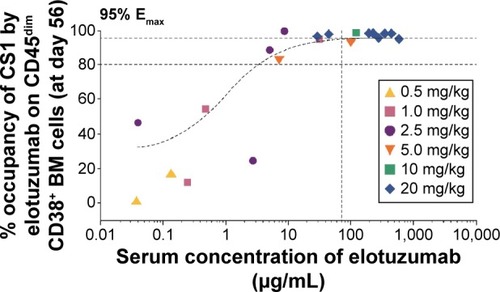
Table 1 Clinical studies with E alone or in combination with established MM therapies
Table 2 Efficacy results of E–L/d versus L/d in treated patients with RRMM (Study ELOQUENT-2)
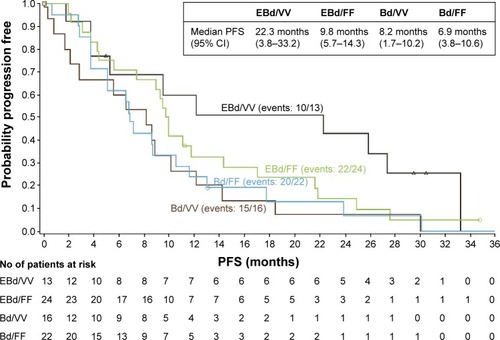
Figure 4 Association of patient FcγRIIIa receptor genotype and PFS in Study CA204-009 (treatment groups: E–B/d vs B/d).
Abbreviations: B, bortezomib; Bd, bortezomib plus dexamethasone; CI, confidence interval; d, dexamethasone; E, elotuzumab; EBd, elotuzumab with bortezomib plus dexamethasone; NK, natural killer; PFS, progression-free survival.