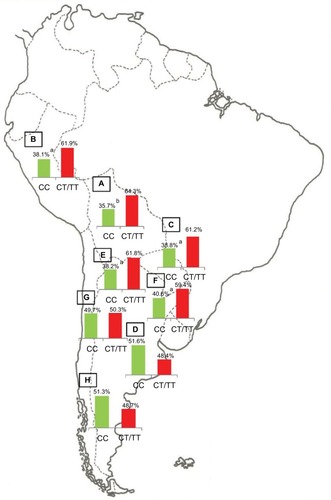
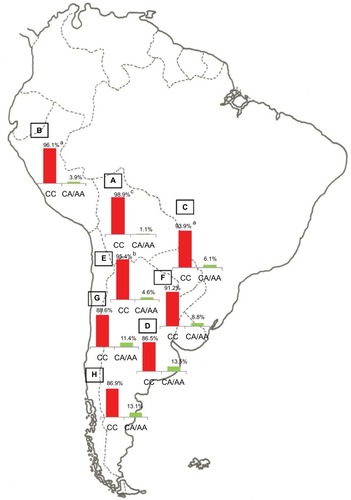
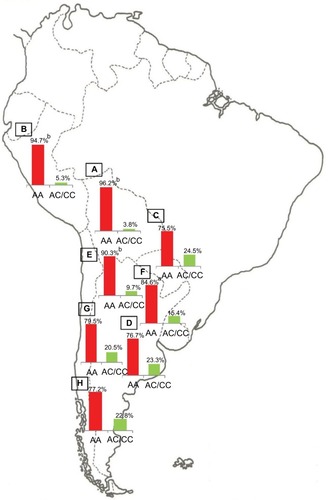
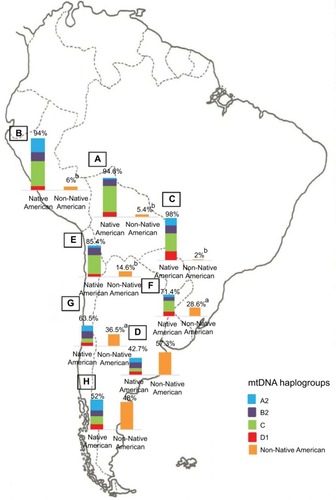
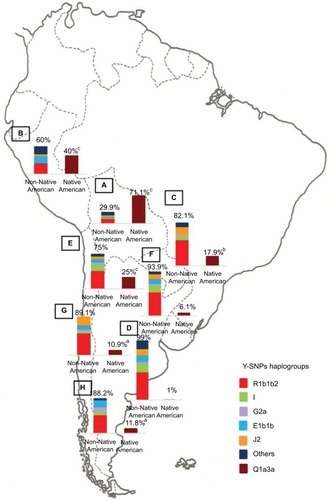
Notes: Prevalence of the Native American and non-Native American Y-SNPs haplogroups in Bolivia (
A, n=185), Peru (
B, n=76), Paraguay (
C, n=98), and the different geographic regions of Argentina: Central (
D, n=991), Northwestern (
E, n=250), Northeastern (
F, n=260), Western (
G, n=175), and Southern (
H, n=115) areas.
ap<0.05 when comparing samples from the Central region of Argentina with those from the Southern and Western regions;
bp<0.01 when comparing samples from the Central region of Argentina with those from Paraguay; and
cp<0.0001 when comparing samples from the Central region of Argentina with those from the Northwestern region, Bolivia, and Peru. Copyright ©2014. Nature Publishing Group. Adapted from Trinks J, Hulaniuk ML, Caputo M, et al. Distribution of genetic polymorphisms associated with hepatitis C virus (HCV) antiviral response in a multiethnic and admixed population.
Pharmacogenomics J. 2014;14(6):549–554.
Citation75 Adapted with permission from Trinks J, Hulaniuk ML, Caputo M, et al. Distribución de polimorfismos de nucleótido simple (SNPs) predictores de la respuesta al tratamiento antiviral en la infección crónica por el virus de la hepatitis C (HCV) en las distintas regiones geográficas de Argentina [Distribution of singlenucleotide polymorphisms (SNPs) predictive of antiviral treatment response in hepatitis C virus (HCV) chronic infection in different geographic regions in Argentina].
Medicina (B Aires). 2014;74 (Suppl 3):S117–S118. Spanish.
Citation76Abbreviation: Y-SNPs, Y-chromosome single-nucleotide polymorphisms.