Figures & data
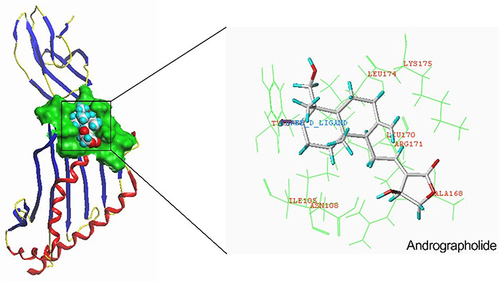
Figure 1 Flow diagram of the study design. (A) The procedure of selecting AZGP1 as the key gene. (B) AZGP1 expression in rectal samples and its visualization; (a) Box plots of AZGP1 differential expression in radioresistant and radiosensitive CRC samples; (b) The integrated SMD and diagnostic meta-analysis of AZGP1 expression in radioresistant versus radiosensitive CRC samples; (c) Survival analysis in patients with CRC in terms of disease-free survival and overall survival. (C) Target prediction for andrographolide and molecular docking. GEO, Gene Expression Omnibus database; SMD, standard mean difference; TCMSP, traditional Chinese medicine systems pharmacology.
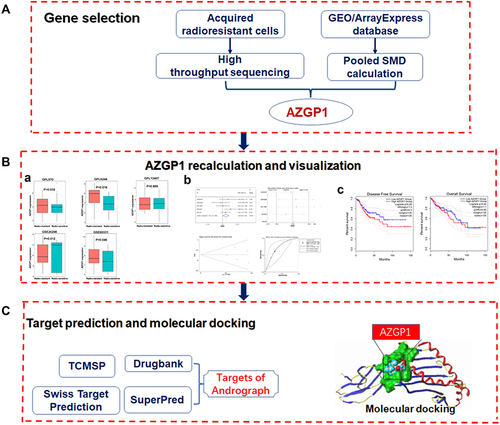
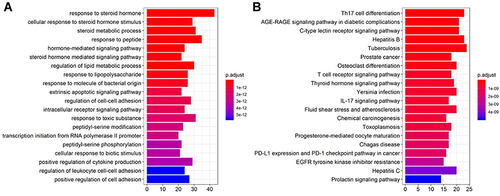
Figure 2 Differentially expressed genes in the acquired radioresistant colorectal cell lines. (A) volcano plots displaying the differentially expressed genes in HCT-116-R cells and HCT-116 cells. (B) volcano plots of the differentially expressed genes in CX-1-R cells versus CX-1 cells. The dark red dots represent the upregulated genes, the deep-blue dots represent the downregulated genes, and the gray dots represent genes with no differential expression. (C) Venn diagrams displaying the upregulated genes in HCT-116-R cells and CX-1-R cells. (D) Venn diagrams displaying the downregulated genes in HCT-116-R cells and CX-1-R cells.
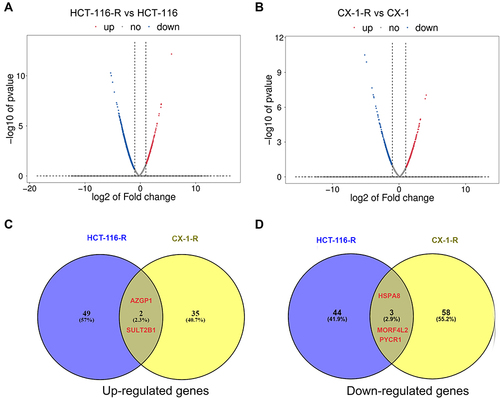
Figure 3 Box plots of AZGP1 differential expression in radioresistant and radiosensitive CRC samples. (A) GPL570 platform containing data from GSE35452 and GSE119409 series; (B) GPL6244 platform containing data from GSE43206 and GSE46862 series; (C) GPL13497 platform containing data from GSE97543 and GSE150082 series; (D) GSE20298 dataset; (E) GSE60331dataset. The Orange boxes represent the expression of AZGP1 in radioresistant samples, while the turquoise boxes represent the expression of AZGP1 in radiosensitive samples.
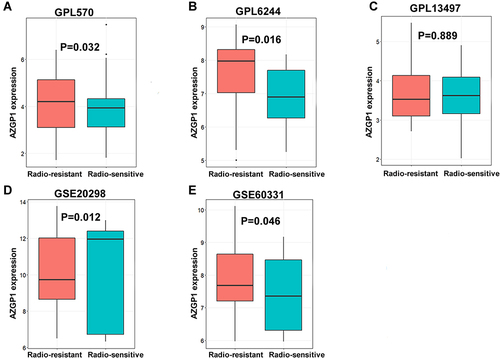
Figure 4 Corresponding ROC curves of AZGP1 differentiated expression in radioresistant versus radiosensitive CRC samples. (A) GPL570 platform containing data from GSE35452 and GSE119409 series; (B) GPL6244 platform containing data from GSE43206 and GSE46862 series; (C) GPL13497 platform containing data from GSE97543 and GSE150082 series; (D) GSE20298 dataset; (E) GSE60331dataset.
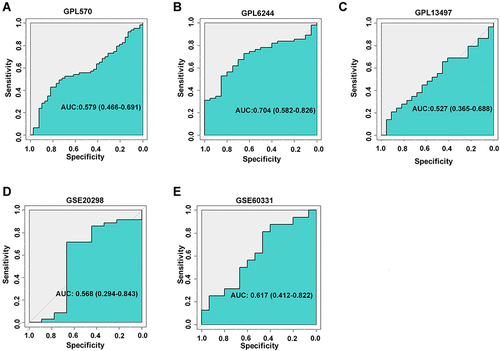
Figure 5 The integrated SMD and diagnostic meta-analysis of AZGP1 expression in radioresistant versus radiosensitive CRC samples. (A) forest plots of SMD values; (B) forest plot of sensitivity analysis; (C) funnel plot; (D) SROC curve of all included studies.
Figure 6 Boxplot of the differentiated expression of AZGP1 in CRC tissue and normal colorectal tissue. (A) GPL96; (B) GPL10558; (C) GPL15207; (D) GSE15781; (E) GSE20842; (F) GSE24713; (G) GSE25071; (H) GSE28000; (I) GSE44076; (J) GSE47063 dataset; (K) GSE87211 dataset; (L) GSE103512 dataset; (M) GSE113513 dataset; (N) GSE115261 dataset; (O) GSE141174 dataset; (P) GSE156355 dataset; (Q) TCGA+CTEx. TCGA, The Cancer Genome Atlas. The Orange boxes represent the expression of AZGP1 in normal colorectal tissue, while the turquoise boxes represent the expression of AZGP1 in CRC tissue.
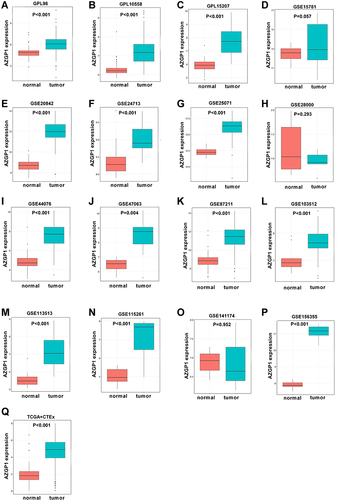
Figure 7 The corresponding ROC curves of AZGP1 differential expression in CRC tissue versus normal colorectal tissue. (A) GPL96; (B) GPL10558; (C) GPL15207; (D) GSE15781 dataset; (E) GSE20842; (F) GSE24713; (G) GSE25071; (H) GSE28000; (I) GSE44076; (J) GSE47063; (K) GSE87211; (L) GSE103512; (M) GSE113513; (N) GSE115261; (O) GSE141174; (P) GSE156355; (Q) TCGA+CTEx. TCGA, The Cancer Genome Atlas.
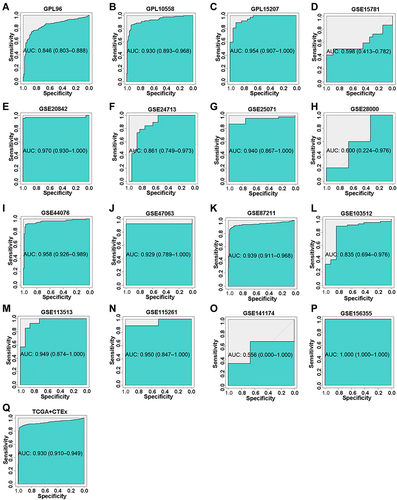
Figure 8 The integrated SMD and diagnostic meta-analysis of AZGP1 expression in CRC tissue versus normal colorectal tissue. (A) forest plots of SMD values; (B) forest plot of sensitivity analysis; (C) funnel plot; (D) SROC curve of the included studies.
Figure 9 AZGP1 expression in normal tissue and colorectal cancer tissue from the Human Protein Atlas database [antibody CAB016087]. (A) Female, age 56. Patient ID 3343. Normal small intestine (T-65000) tissue, NOS(M-00100), low expression of AZGP1. The relative results of glandular cells are as follows: staining: low; intensity: moderate; quantity: <25%; location: cytoplasmic/membranous. (B) Female, age 56. Patient ID 1423. Normal colon (T-67000) tissue, NOS(M-0010), deficient expression of AZGP1. The relative results of glandular cells are as follows: staining: not detected; intensity: negative; quantity: none; location: none. (C) Female, age 66. Patient ID 2060. Normal rectum (T-68000) tissue, NOS(M-00100), deficient expression of AZGP1. The relative results of glandular cells are as follows: staining: not detected; intensity: negative; quantity: none; location: none. (D) Male, age 51. Patient ID 3298. Rectum (T-68000) adenocarcinoma, NOS(M-81403), moderate expression of AZGP1. The relative results of tumor cells are as follows: staining: medium; intensity: moderate; quantity: 75%–25%; location: cytoplasmic/membranous.
![Figure 9 AZGP1 expression in normal tissue and colorectal cancer tissue from the Human Protein Atlas database [antibody CAB016087]. (A) Female, age 56. Patient ID 3343. Normal small intestine (T-65000) tissue, NOS(M-00100), low expression of AZGP1. The relative results of glandular cells are as follows: staining: low; intensity: moderate; quantity: <25%; location: cytoplasmic/membranous. (B) Female, age 56. Patient ID 1423. Normal colon (T-67000) tissue, NOS(M-0010), deficient expression of AZGP1. The relative results of glandular cells are as follows: staining: not detected; intensity: negative; quantity: none; location: none. (C) Female, age 66. Patient ID 2060. Normal rectum (T-68000) tissue, NOS(M-00100), deficient expression of AZGP1. The relative results of glandular cells are as follows: staining: not detected; intensity: negative; quantity: none; location: none. (D) Male, age 51. Patient ID 3298. Rectum (T-68000) adenocarcinoma, NOS(M-81403), moderate expression of AZGP1. The relative results of tumor cells are as follows: staining: medium; intensity: moderate; quantity: 75%–25%; location: cytoplasmic/membranous.](/cms/asset/11976def-d6a6-4e15-9384-0571b4af6360/dpgp_a_12153416_f0009_c.jpg)
Figure 10 Survival analysis in patients with CRC in terms of disease-free survival and overall survival. (A) Kaplan-Meier curves for disease-free survival; (B) Kaplan-Meier curves for overall survival.
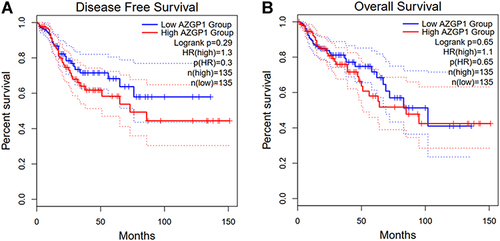
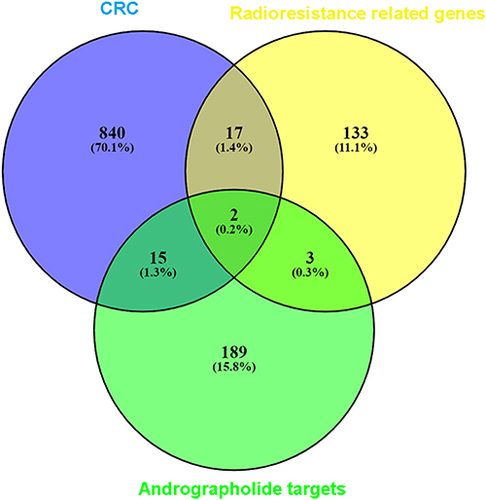
Figure 11 Venn diagrams for the intersections between genes acting on andrographolide and radiotherapy-resistance related genes in colorectal cancer. The 209 andrographolide targets from the different databases and text mining, and the overlapping targets were AZGP1 and SULT2B1.