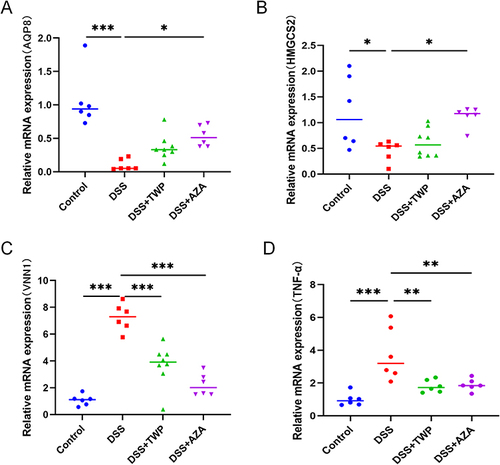
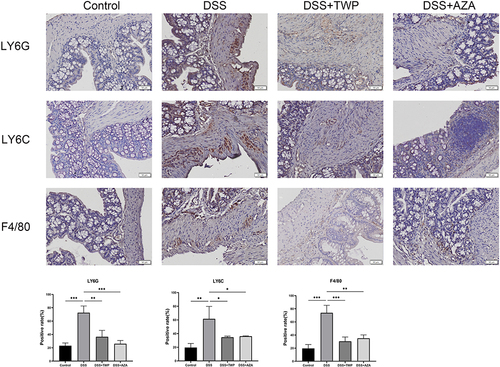
Figures & data
Figure 1 (A) Volcano map of DEGs in GSE87466 and GSE179285, red dots represent the upregulated genes and the green dots represent the downregulated genes; (B) Heat map of DEGs.
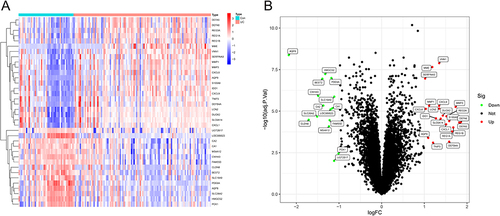
Figure 2 (A) Results of GO enrichment analysis; (B) results of KEGG enrichment analysis; (C) results of DO analysis; (D) GSEA analysis results displaying the top 5 signaling pathways.
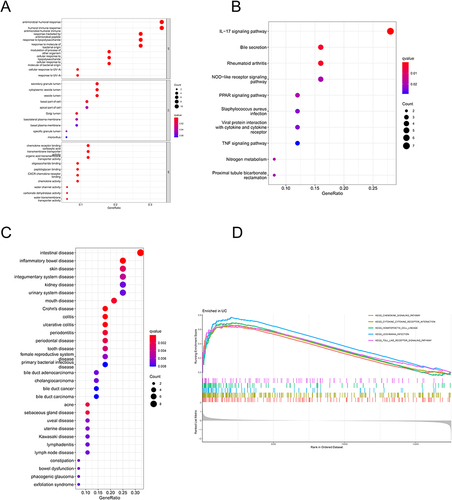
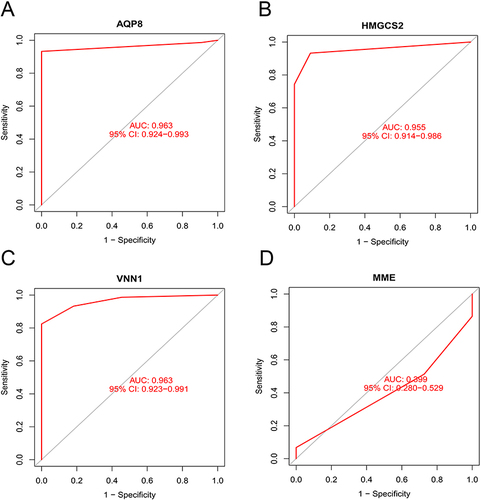
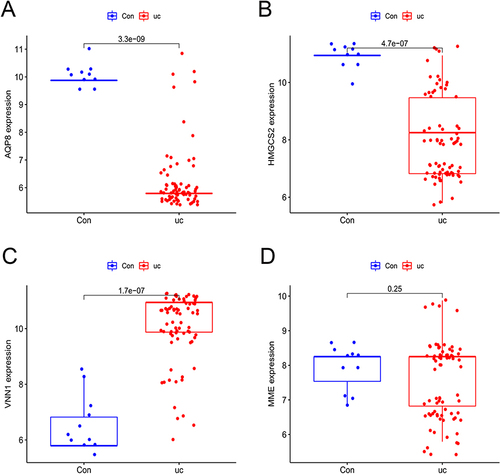
Figure 3 (A) LASSO logistic regression algorithm used to analyze the feature genes; (B) SVM-RFE algorithm used to analyze the feature genes; (C) Venn diagram of the intersection of two sets of feature genes.
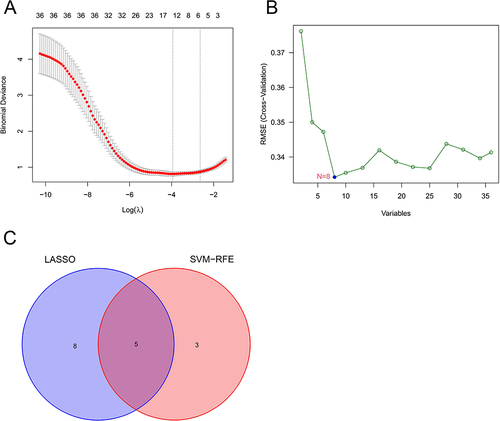
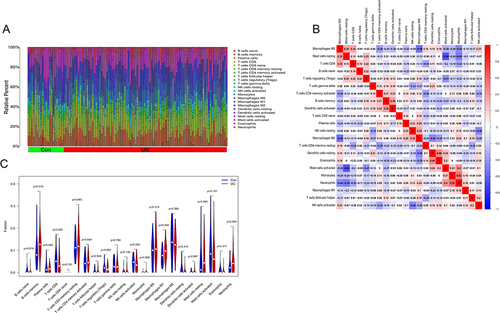
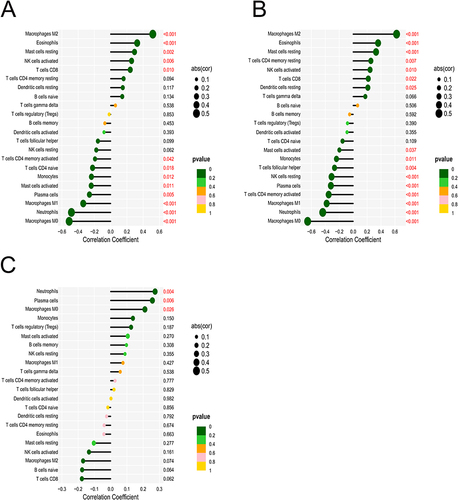
Figure 6 (A) Results of immune cell infiltration in the colon of UC patients and normal subjects; (B) heat map of the correlation of 22 immune cell species in UC; (C) the violin plot of the ratio of 22 immune cell species.