Figures & data
Table 1 Molecular Characterization of Deafness-Associated Mt-tRNAGlu Mutations
Figure 2 PCR amplification of mt-tRNAGlu gene in subjects with NSHL, arrow indicated the PCR product, which was 1050-bp.
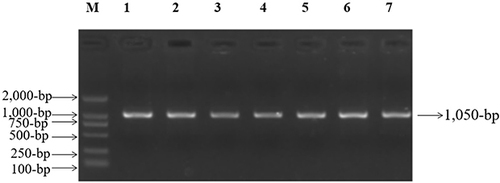
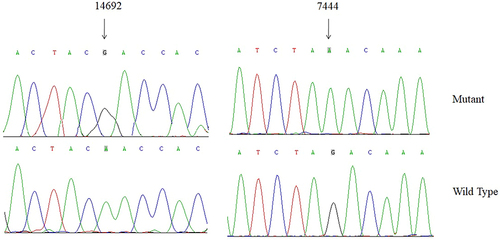
Figure 3 Sequence alignment of mt-tRNAGlu gene from different species, arrows indicated the positions of 37, 54, 55 and 64, corresponding to the m.T14709C, m.A14693G, m.A14692G and m.A14683G variants.
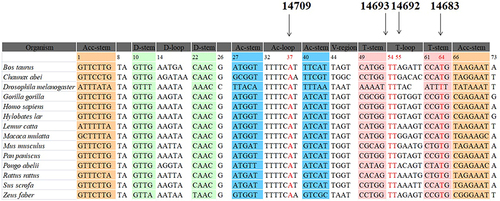
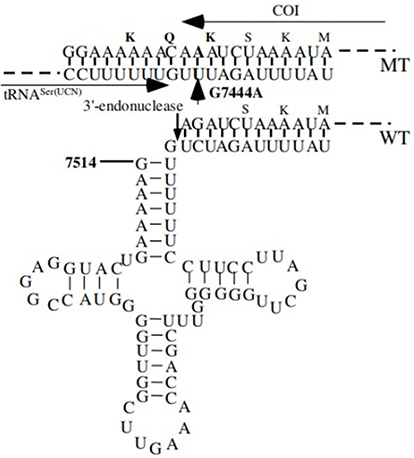
Figure 4 Cloverleaf structure of mt-tRNAGlu gene, arrows indicated the positions of m.T14709C, m.A14693G, m.A14692G and m.A14683G variants.
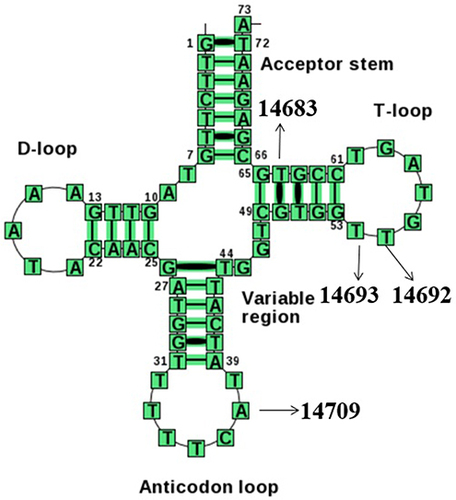
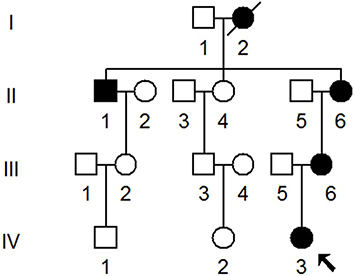
Table 2 Summary of Clinical and Molecular Data for Several Members of the Chinese Family
Figure 5 Audiological examination of matrilineal relatives of one pedigree with NSHL, (X) left ear; (O) right ear.
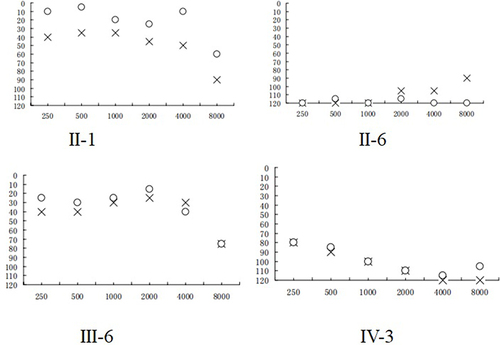
Table 3 mtDNA Variants in One Chinese Family with Hearing Impairment
Data Sharing Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request (Yu Ding: [email protected]).