Figures & data
Figure 1 Identification of differentially expressed genes.
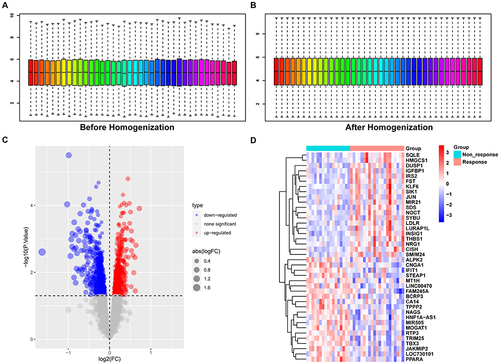
Figure 2 Enrichment analysis of DEGs.
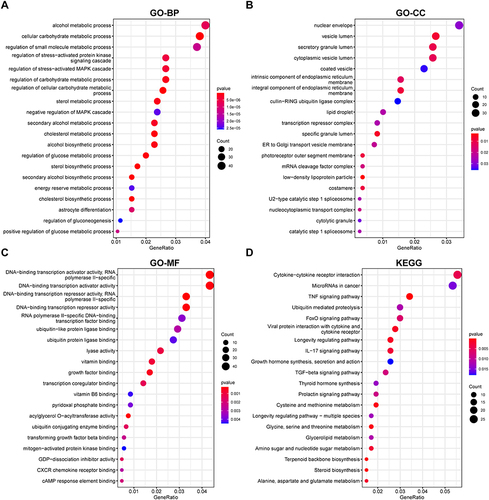
Figure 3 Identification of glycosylation-related DEGs.
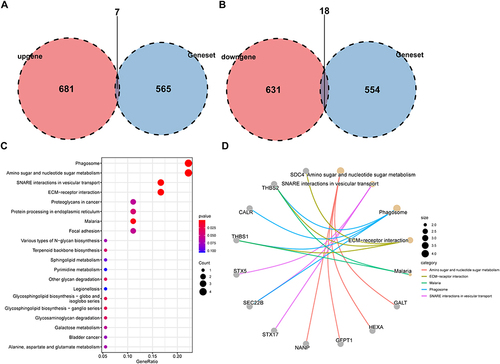
Figure 4 Exhibition of glycosylation-related DEGs.
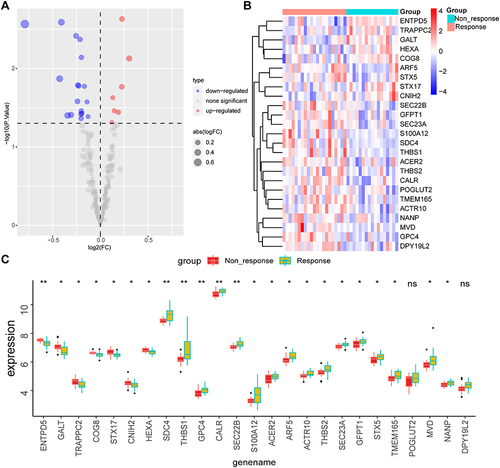
Figure 5 PPI network for DEGs.
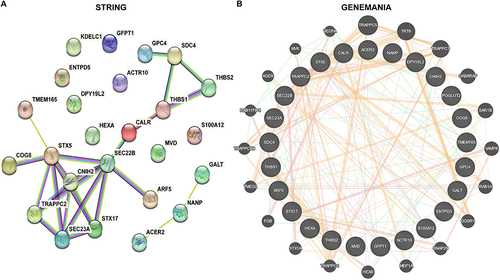
Figure 6 Selection of hub genes via machine learning.
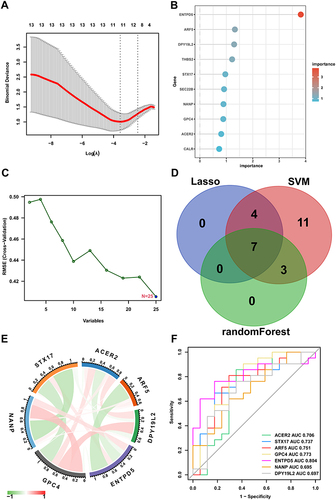
Figure 7 The correlation analysis of immune cell infiltration.
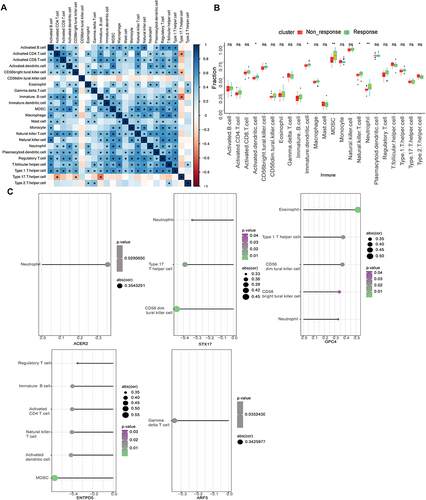
Figure 8 The Co-expression analysis of hub genes.
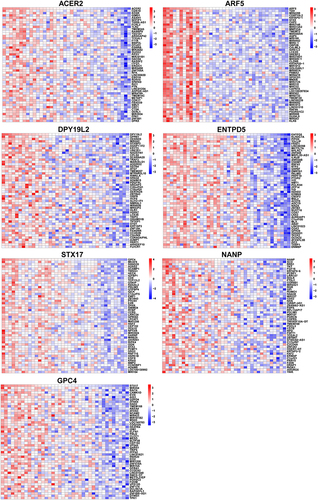
Figure 9 GSEA of hub genes.
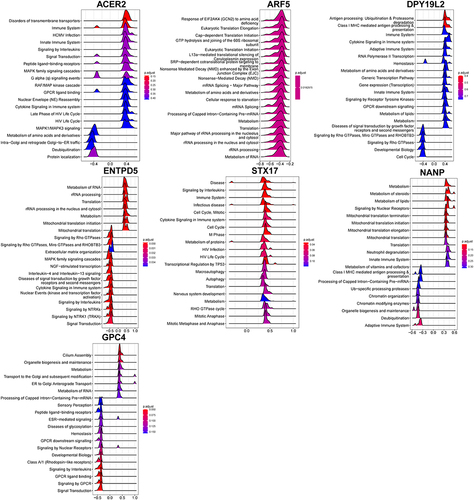
Figure 10 Establishment of the miRNA-TF-genes network.
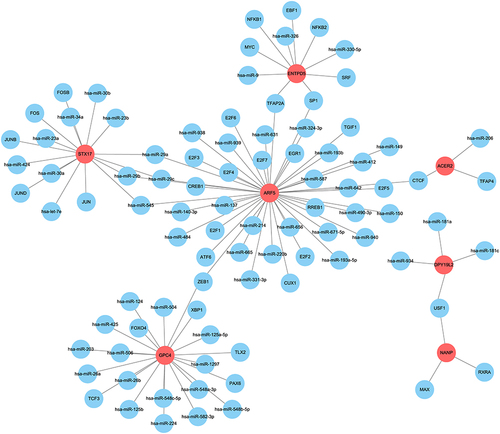
Figure 11 Validation of glycosylation-related hub genes.
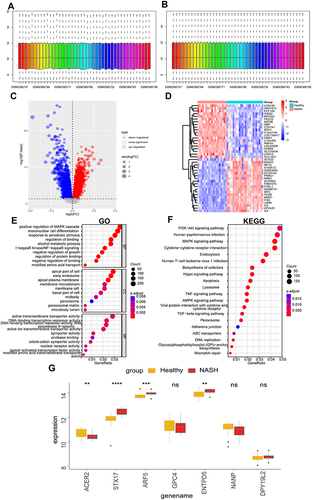
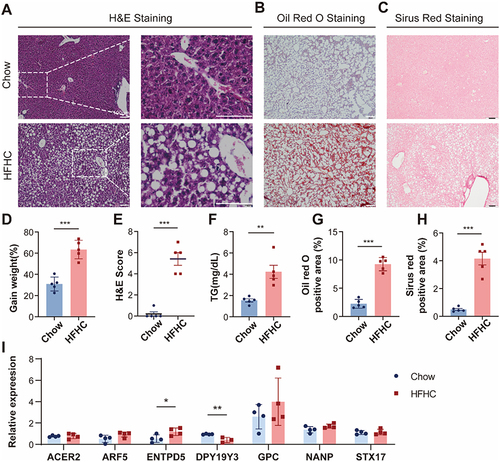
Figure 12 Validation of hub genes in HFHC mice.
Data Sharing Statement
Raw data of this study are available from the corresponding author Xiujun Cai upon request.
