Figures & data
Figure 1 Representative images of organization of SEPT9 filaments in mammalian cells at different stages of the cell cycle.
Abbreviation: SUE, septin-unique element.
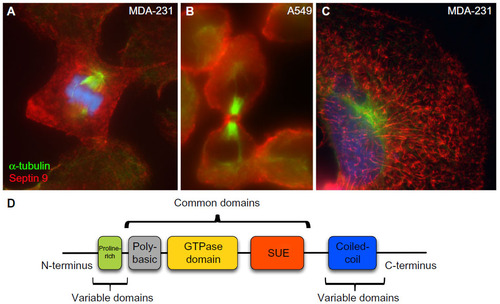
Table 1 Chromosomal mapping and transcriptional variants of Septin family members
Figure 2 Representation of the crystal structure of the SEPT7, SEPT2, and SEPT2 complex.
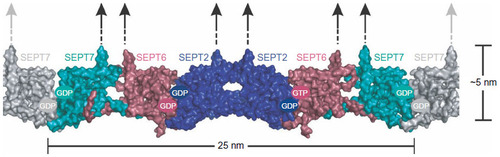
Figure 3 Plotting of frequency (y-axis) of copy-number alterations (CNAs) observed in cancer patients analyzed as part of the Cancer Genome Atlas (TCGA).
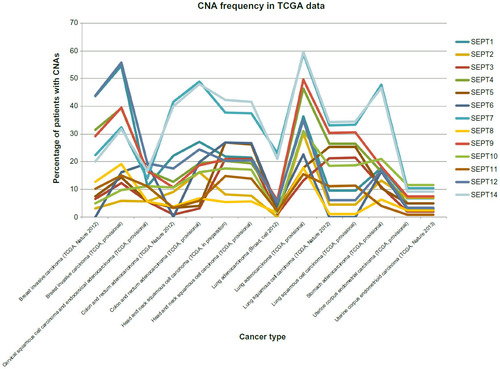
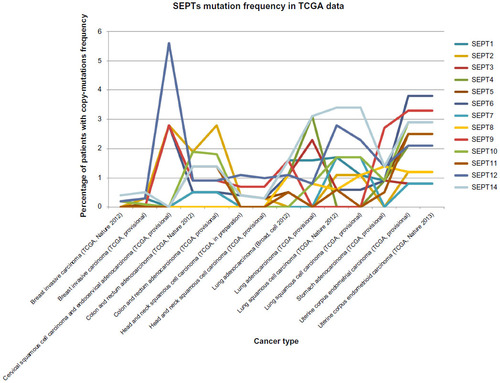
Figure 4 Plotting of mutation frequency (y-axis) observed in cancer patients analyzed as part of the Cancer Genome Atlas (TCGA).