Figures & data
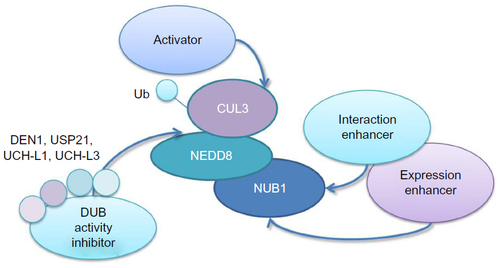
Figure 1 Domain information of NUB1/NUB1L.
Notes: Numbers indicate the serial number of amino acids. Top: NUB1. Bottom: NUB1L. NUB1 has one UbL domain (Ile85 to Val147), two CC (36–67 aa, 155–203 aa), two UBA domains (Asp376 to Asn413, Ser477 to His514), one Cys box-like sequence (Lys306 to Glu320), one His box-like sequence (Tyr343 to Tyr362), a bipartite NLS (Arg414 to Arg431), and a PEST sequence (His514 to His568). NUB1L has a 14-amino-acid insertion between 451 and 452 residues in the protein sequence of NUB1, thus having an extra UBA domain (UBA2, 432–469 aa).
Abbreviations: aa, amino acids; CC, coiled coil regions; Cys, Cys-box-like sequence; His, His-box-like sequence; NLS, nuclear localization sequence; NUB1, negative regulator of ubiquitin-like proteins 1; NUB1L, negative regulator of ubiquitin-like proteins 1 long; PEST, one of the signals of degradation enriched with proline, glutamate, serine, and threonine; UbL, ubiquitin-like; UBA, ubiquitin-associated.
Abbreviations: aa, amino acids; CC, coiled coil regions; Cys, Cys-box-like sequence; His, His-box-like sequence; NLS, nuclear localization sequence; NUB1, negative regulator of ubiquitin-like proteins 1; NUB1L, negative regulator of ubiquitin-like proteins 1 long; PEST, one of the signals of degradation enriched with proline, glutamate, serine, and threonine; UbL, ubiquitin-like; UBA, ubiquitin-associated.
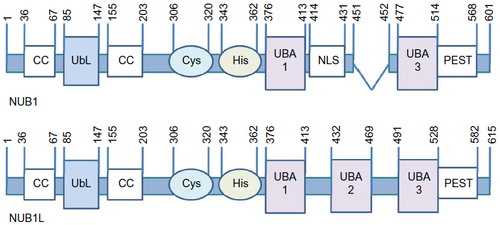
Figure 2 Possible mechanism models of NUB1-mediated mHTT clearance.
Notes: NUB1 may function as a bridging protein between HTT and neddylated CUL3-based E3 ligases via physical interaction with HTT and NEDD8. CUL3 interacts with ROC1/Rbx1 which recruits charged ubiquitin E2s into the complex when activated by neddylation of NEDD8 Gly-76 on its lysine residue, promoting the ubiquitin–proteasomal degradation of both mHTT and wtHTT in the presence of mHTT, with KLHL22 as the substrate receptor protein of CUL3 for HTT; or CUL3 may be indirectly involved via regulating other NUB1 interactors. Meanwhile, there is a potential feedback relationship between mHTT and NUB1, in which mHTT may have a compensatory way to elevate NUB1 expression and NUB1 functioning might need activation by mHTT, possibly via enhancing interaction with other proteins in the pathway.
Abbreviations: CUL3, cullin3; KLHL22, kelch-like family member 22; mHTT, mutant huntingtin protein; wtHTT, wild type huntingtin protein; N17, the first 17 amino acids in the N-terminus of HTT; KKK, the three lysine residues in N17; NEDD8, neural precursor cell expressed, developmentally downregulated 8; NUB1, negative regulator of ubiquitin-like proteins 1; polyQ, polyglutamine; Rbx1, RING-box protein 1; ROC1, regulator of cullins 1; Ub, ubiquitin.
Abbreviations: CUL3, cullin3; KLHL22, kelch-like family member 22; mHTT, mutant huntingtin protein; wtHTT, wild type huntingtin protein; N17, the first 17 amino acids in the N-terminus of HTT; KKK, the three lysine residues in N17; NEDD8, neural precursor cell expressed, developmentally downregulated 8; NUB1, negative regulator of ubiquitin-like proteins 1; polyQ, polyglutamine; Rbx1, RING-box protein 1; ROC1, regulator of cullins 1; Ub, ubiquitin.
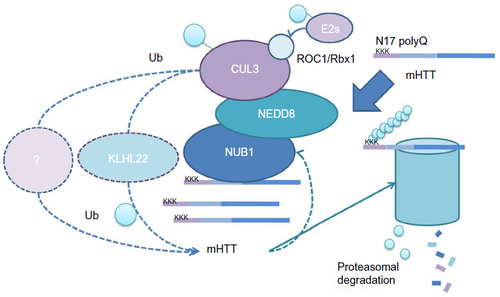
Table 1 Targeting strategies for NUB1
Figure 3 Targeting strategies for NUB1.
Notes: We have suggested several approaches for drug discovery of NUB1-mediated mHTT clearance: 1) elevating NUB1 expression by NUB1 transcriptional enhancers; 2) enhancing NUB1 function by enhancers of NUB1 interaction with key proteins in the pathway; 3) blocking deneddylation of CUL3 by activity inhibitors of NEDD8 DUBs; and 4) enhancing enzymatic activity of CUL3-based E3 ligases by direct activators.
Abbreviations: CUL3, cullin3; DUB, deubiquitinating enzyme; DEN1, deneddylase 1; NEDD8, neural precursor cell expressed, developmentally downregulated 8; NUB1, negative regulator of ubiquitin-like proteins 1; Ub, ubiquitin; UCH-L1, ubiquitin C-terminal hydrolase 1; UCH-L3, ubiquitin C-terminal hydrolase 3; USP21, ubiquitin-specific peptidase 21; mHTT, mutant huntingtin protein.
Abbreviations: CUL3, cullin3; DUB, deubiquitinating enzyme; DEN1, deneddylase 1; NEDD8, neural precursor cell expressed, developmentally downregulated 8; NUB1, negative regulator of ubiquitin-like proteins 1; Ub, ubiquitin; UCH-L1, ubiquitin C-terminal hydrolase 1; UCH-L3, ubiquitin C-terminal hydrolase 3; USP21, ubiquitin-specific peptidase 21; mHTT, mutant huntingtin protein.