Figures & data
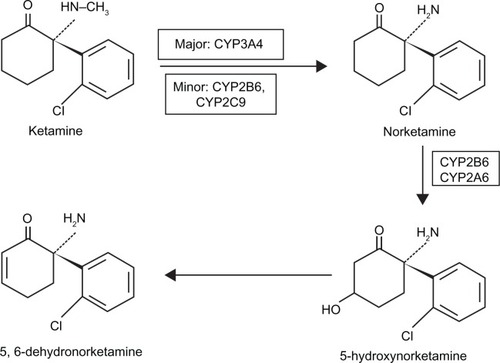
Figure 1 Schematic diagram of NMDA receptor complex. The NMDA receptor is an ionotropic glutamate receptor for controlling synaptic plasticity and memory function.Citation47,Citation49 Glutamate (and NMDA) binds to the agonist site on the NMDA receptors. PCP, ketamine, and dizocilpine bind to the PCP receptor in the inside of the NMDA receptors. Glycine and D-serine bind to a glycine modulatory site on the NMDA receptors. The NMDA receptor is blocked by Mg2+ in a voltage sensitive manner. Activation of NMDA receptor by binding of both glutamate and glycine results in the opening of the channel. This allows voltage-dependent flow of Na+ and small amounts of Ca2+ ions into the cell and K+ out of the cell. The symbol (−) denotes inhibitory effect.
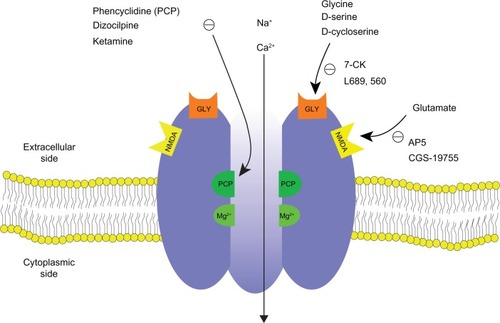
Figure 2 The pathway of ketamine metabolism in phase I. Ketamine is N-demethylated by liver cytochrome P450 enzymes (major: CYP3A4; minor: CYP2B6 and CYP2C9) into norketamine.Citation91,Citation92 CYP2B6 and CYP2A6 have been identified to hydroxylate norketamine into 5-hydroxynorketamine.Citation91 The chemical structures are depicted for S-ketamine and its metabolites.