Figures & data
Figure 1 Strategy for detecting and distinguishing homozygous/heterozygous genotypes of the HLA-C*03:02 allele. (A) PCR procedures: Step 1. The primer set HLACB1F/HLACB1R specifically amplified the exon 2–3 sequence of the HLA-C gene. Step 2. The 912 bp PCR product from step 1 was then used as a template for the step 2 PCR reactions, which used three primer sets. (B) Different patterns can be obtained with the three primer sets in the second PCR step according to the HLA-C*03:02 zygosity. (*): allele number.
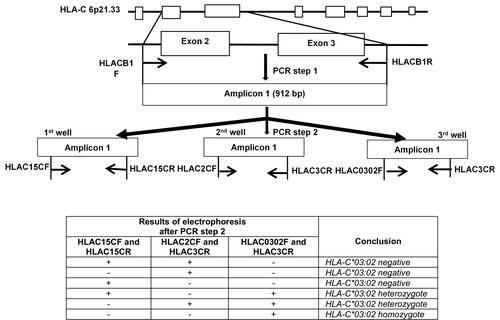
Table 1 Sequences of Primer Sets Used for the Two PCR Steps
Figure 2 Binding sites of the primer set used in the step 1 PCR. (A) Forward primer HLACB1F: a mismatch (replacement of G with T) at the penultimate position of the 3′ terminus is shown in grey. (B) Reverse primer HLACB1R. The reference sequences were obtained from https://www.ebi.ac.uk/ipd/imgt/hla/.Citation17
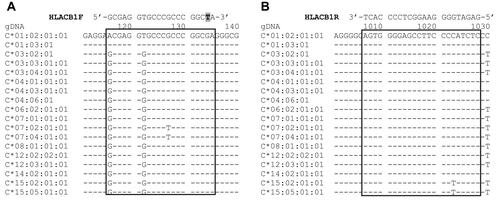
Figure 3 Binding sites of the primer sets used in the step 2 PCR. (A) HLAC0302F has one mismatch (replacement of C with T) at the penultimate position of the 3′ terminus; HLAC2CF has one mismatch (replacement of T with C) at the third position from the 3′ terminus. The mismatches are highlighted in grey; (B) HLAC3CR and HLAC15CR have two different nucleotides (highlighted in grey) at the 3′ terminus that ensure the specificity of the primers. The reference sequences were obtained from https://www.ebi.ac.uk/ipd/imgt/hla/.Citation17

Table 2 AS-PCR Results of 10 Samples of Known Genotypes
Figure 4 The detection of the HLA-C*03:02 allele in 10 samples of known genotype. (A) Step 1: HLA-C exon 2–3 amplicon, 912 bp; (B) Step 2: amplicon from the primer set HLAC15CF and HLAC15CR, 569 bp; (C) Step 2 amplicon from the primer set HLAC2CF and HLAC3CR, 241 bp; (D) Step 2: amplicon from the primer set HLAC0302F and HLAC3CR, 241 bp. (*): allele number.
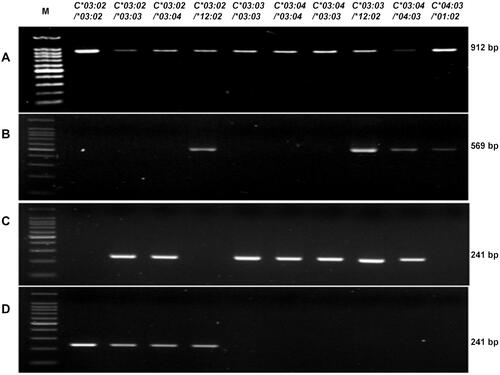
Table 3 Comparison of the AS-PCR Method with PCR-SBT
Table 4 HLA-C*03:02 Distributions in Vietnamese Kinh People
