Figures & data
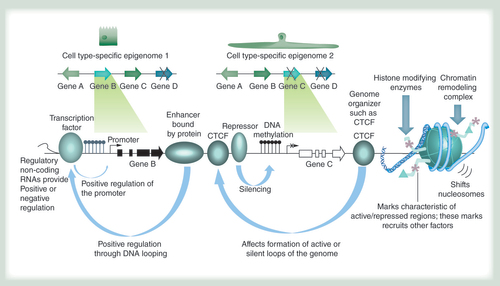
The epigenome allows cell types with identical genomes (schematically depicted at the top) to exhibit differential gene expression patterns through a variety of mechanisms. They include, from left to right: regulatory RNAs of different types such as long noncoding RNAs, miRNAs and others; recruitment of transcription factors that can activate gene expression; enhancer-binding proteins that can activate gene expression by looping to promoters in cis or in trans (when different chromosomes interact); proteins like CTCF that mediate looping interactions in concert with other proteins such as cohesin and thereby support the three-dimensional organization of the genome; the binding of repressor proteins that can silence genes; DNA methylation that is usually absent in active regulatory regions such as promoters and enhancers (hydroxymethylation, thought to occur during DNA methylation removal, and other modifications can also occur); a variety of histone modifying enzymes that lead to protein interactions associated with different chromatin states; nucleosome remodeling complexes that can lead to the removal or deposition of nucleosomes in active and repressed genomic regions, respectively.
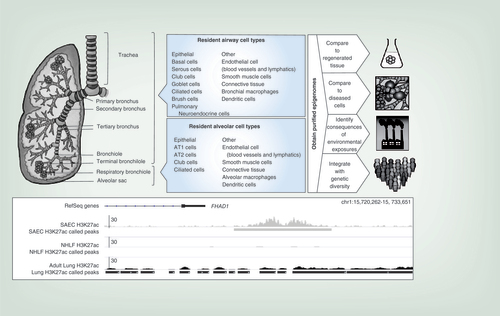
Only the right lung is shown. Airway and alveolar compartments and their cell types are indicated. The composition of human respiratory bronchioles has been poorly investigated but is thought to contain club cells and rare ciliated cells. At right, different applications for obtained epigenomes are indicated. At the bottom, an example is shown of epigenetic differences between lung cell types: histone 3 lysine 27 acetylation (H3K27Ac, a mark indicating an active enhancer) ChIP-seq data is compared between small airway epithelium, normal human lung fibroblasts and adult lung tissue. The presence of an enhancer is noted downstream of the FHAD gene in small airway epithelial cells but not in lung fibroblasts; the element is not clearly detectable in adult lung, which is a mix of many different cell types.