Figures & data
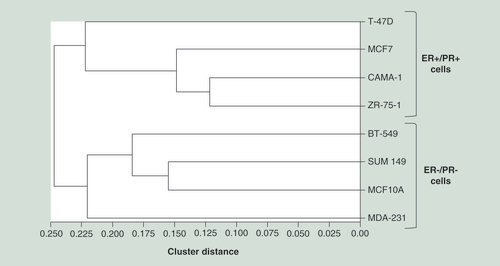
Relative mRNA expression of human DNMT3B2 (gray bar) and its alternative spliced isoform DNMT3B3 (black bar). Results are expressed in mRNA copy number corrected with the reference gene HPRT1 as mean ± SEM; n = 4.
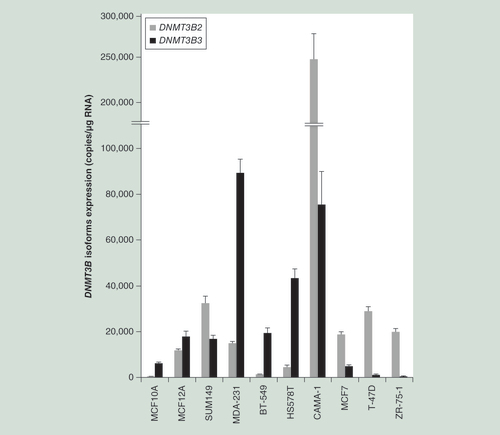
Hierarchical clustering of parental (untransfected) cell lines based on genome-wide methylation data of each cell line. This analysis was performed with GenomeStudio calculating dissimilarities based on absolute correlation using a 1-r distance measure. Rows represent each parental (untransfected) cell line.