Figures & data
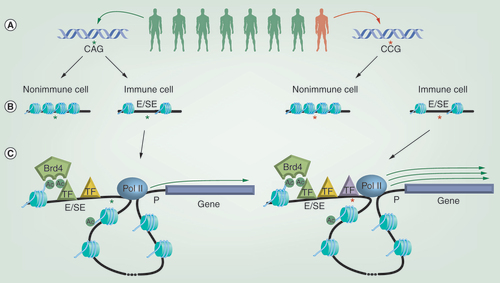
(A) Genome-wide association studies have shown that individuals harboring specific rare single nucleotide polymorphisms (SNPs) in their genetic sequence can show an increased incidence of developing particular immune-mediated diseases. (B) Rather than occurring in the protein-coding region of genes, most disease-associated SNPs sit within regions of the genome that regulate gene expression, such as enhancers (Es) and super enhancers (SEs), which are characterized by possessing relatively open and nucleosome free chromatin structures. SNPs associated with immune diseases are enriched within Es/SEs found selectively within relevant immune cell types. (C) Es/SEs are able to regulate the expression of distal genes through the ‘looping out’ of intervening DNA, bringing the E/SE in close proximity to the gene promoter (P). E/SE function is linked to the recruitment of transcription factors (TFs) and epigenetic regulators, such as Brd4, which binds to acetylated histones and TFs via its bromodomains. One mechanism by which SNPs may alter gene expression is by changing the binding sites for TFs within Es/SEs and ultimately the efficiency of recruiting components of the transcriptional machinery, such as RNA polymerase II. In the example depicted, the disease-associated SNP leads to increased TF recruitment and elevated expression of the gene controlled by that E/SE.