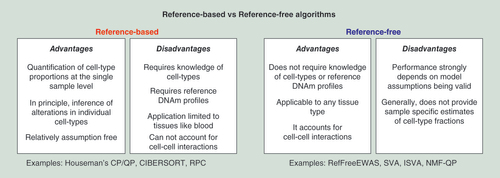
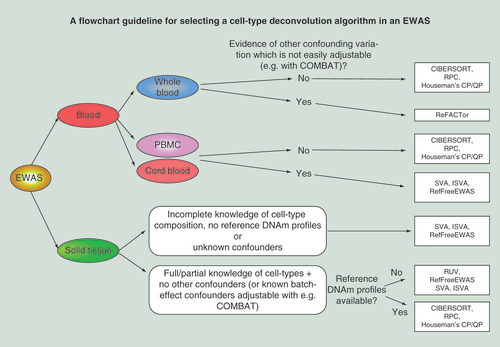
Figures & data
Table 1. Algorithm name, whether it is reference-free, reference-based or semireference free, what inference is possible with the algorithm, tissue types on which it has been successfully applied, whether algorithm adjusts for confounders other than cell-type composition, programming language in which it is available, website link and main reference.