Figures & data
Table 1. First- and second-generation participant characteristics and recruitment source (n = 62).
Table 2. Alcohol dependence and smoking status of second-generation parents of third generation.
Table 3. Number of relatives with alcohol dependence across generations.
Table 4. Number of parents and grandparents with alcohol dependence.
Table 5. Third generation participant characteristics and recruitment source (n = 441).
Table 6. Personal use of substances prior to DNA collection for third generation participants (n = 441).
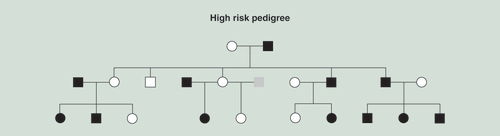
Black squares or circles indicate the presence of an alcohol dependence diagnosis. Gray indicates phenotype unknown. Squares indicate males; circles females.
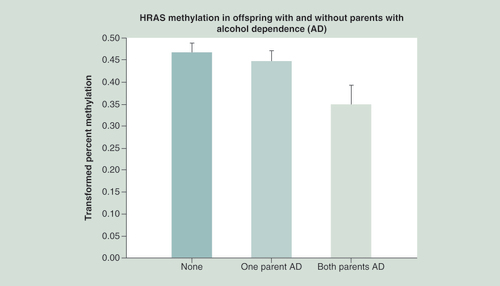
Statistical analysis controlled for the age that DNA was obtained from offspring, prenatal use of substances by mothers (alcohol, drugs and cigarettes), and the personal use by the offspring up to the time that DNA was obtained. Hypomethylation of HRAS in offspring is seen in association with increased loading of parental AD (p = 0.022). This hypomethylation suggests increased expression of the oncogene.
AD: Alcohol dependence.
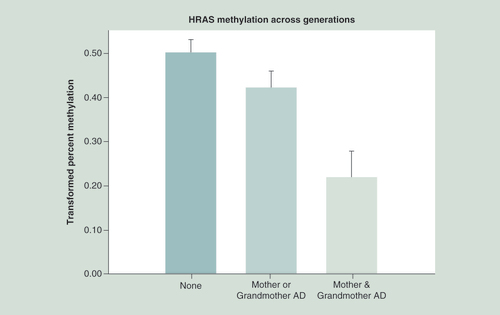
Statistical analysis controlled for age at DNA collection, the mothers’ prenatal use of substances and the offspring’s personal use up to the time of DNA collection. Hypomethylation of HRAS CpG sites is seen in association with increased familial AD across generations (p = 0.003).
AD: Alcohol dependence.
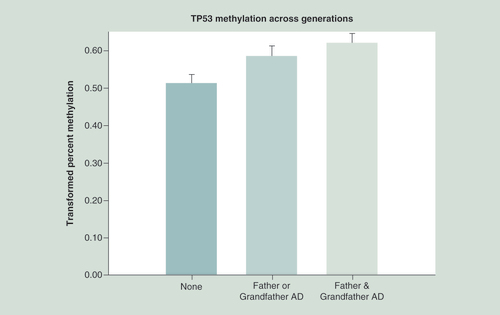
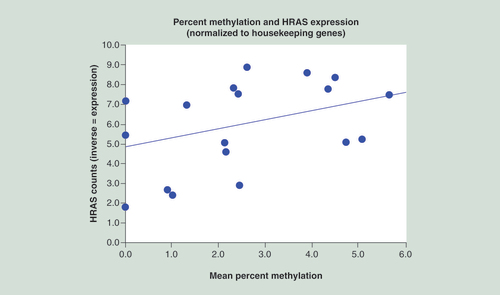
The parametric relationship showed an r of 0.38 with a marginally significant value, with the ordinal relationship showing good significance (p = 0.001).
TP53 methylation in offspring was compared with those without a father or grandfather with alcohol dependence, with those with either a father or grandfather and those having both. Statistical analysis controlled for age at DNA collection, the mothers’ prenatal use of substances and the offspring’s personal use up to the time of DNA collection. Hypermethylation of CpG sites is seen in association with increased familial AD across generations (p = 0.0068). This hypermethylation suggests lesser expression of the tumor suppressor gene.
AD: Alcohol dependence.