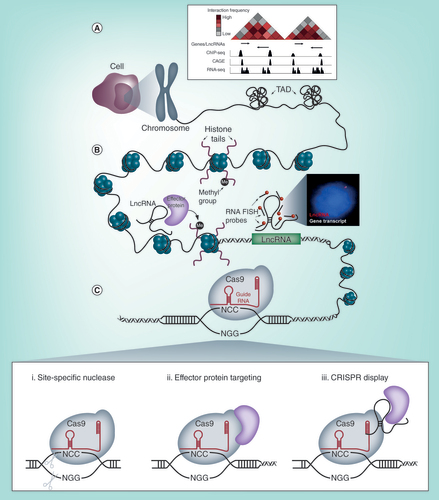
Figures & data
Table 1.
Summary of the emerging methods used to study long noncoding RNA biology.