Figures & data
Table 1. Clinical description of cohort.
Table 2. Oligonucleotide sequences.
Table 3. Differentially methylated probes identified by Minfi.
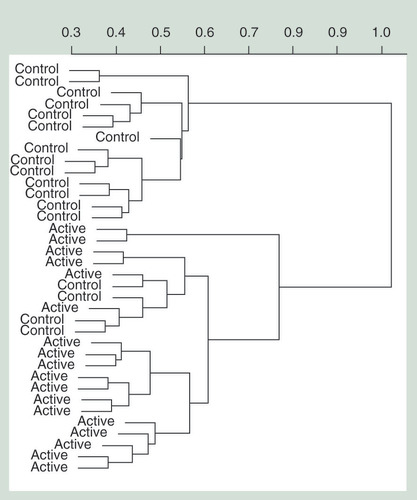
Hierarchical clustering performed using the top 100 most significant probes (Supplementary Table 3). The change in relative methylation values (β) is represented on the y-axis and individuals from each treatment are represented on the x-axis.
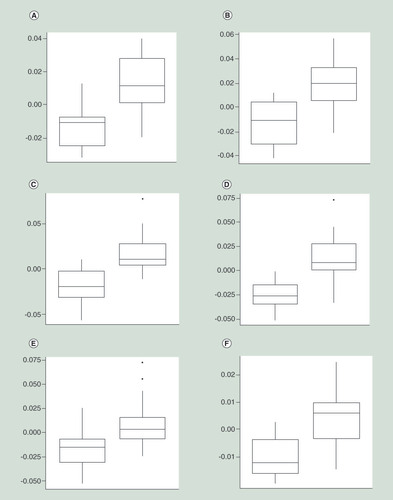
Change in methylation values is represented on the y-axis, in each instance the boxplot on the left represents the control group and the boxplot on the right represents the treatment group. (A) Chr1:204985932; (B) Chr3:183913285; (C) Chr8:25829073; (D) Chr11: 16430764; (E) Chr17: 39120012; (F) Chr10:8092775.