Figures & data
Table 1. Epigenetic regulation in cancer.
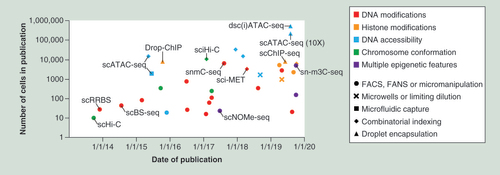
The publication dates and cell numbers are graphed for single-cell epigenomic methods, with the names of key methods shown. The epigenetic features assessed are indicated by colour and the means of cell isolation are indicated by symbol. This field has seen substantial growth in the diversity of epigenetic modifications that can be studied in single cells and significant improvements to through-put driven by combinatorial indexing and droplet encapsulation. Supplementary Table 1 provides further details.
FACS: Fluorescence activated cell sorting; FANS: Fluorescence activated nuclei sorting.
Cellular heterogeneity in cancer is associated with clinical challenges such as metastasis, therapeutic response and relapse. Single-cell technologies will identify target cell populations (by dimensional reduction methods) that could not be distinguished using traditional techniques. Single-cell epigenomics will identify modifications specific to those cells. For example, DNA methylation at a certain allele may be observed only in therapy resistant cells (red flag). Based on this information, simplified tests will be developed for clinical use. For example, amplicon bisulphite sequencing of specific loci may be used to advise clinicians of the proportion of cells in a sample with therapeutic resistance (each line indicates a sequencing read, black and white circles represent methylated and unmethylated cytosines respectively, the methylation pattern associated with therapy resistance is highlighted in red).