Figures & data
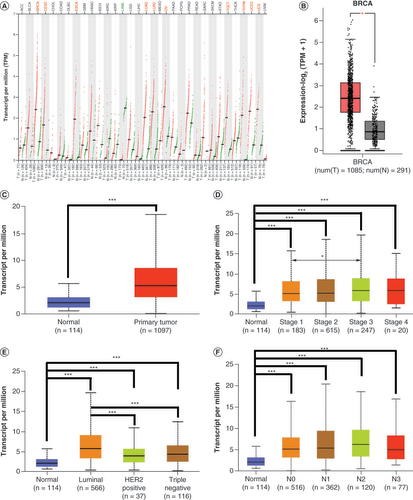
(A) Expression pattern of SIX4 in different types of malignancies. Dots in the figure represent different samples: red for tumors and green for normal tissue. In different types of malignancies, the cancerous tissues with significantly increased SIX4 are labeled in red and include BRCA, CESC, ESCA, LUAD, LUSC, OV, TGCT, THYM, UCEC and UCS, while malignant tissues with decreased SIX4 are labeled in green and include LAML. (B & C) The increased SIX4 level in breast cancer compared with normal tissues in GEPIA and UALCAN. (D) Expression of SIX4 in different stages of breast cancer. (E) Expression of SIX4 in different molecular subtypes of breast cancer. (F) Expression of SIX4 in different lymph node statuses.
BRCA: Breast invasive carcinoma; CESC: Cervical squamous cell carcinoma and endocervical adenocarcinoma; ESCA: Esophageal carcinoma; GEPIA: Gene Expression Profiling Interactive Analysis; LAML: Acute myeloid leukemia; LUAD: Lung adenocarcinoma; LUSC: Lung squamous cell carcinoma; OV: Ovarian serous cystadenocarcinoma; TGCT: Testicular germ cell tumor; THYM: Thymoma; UALCAN: University of Alabama at Birmingham Cancer portal; UCEC: Uterine corpus endometrial carcinoma; UCS: Uterine carcinosarcoma.
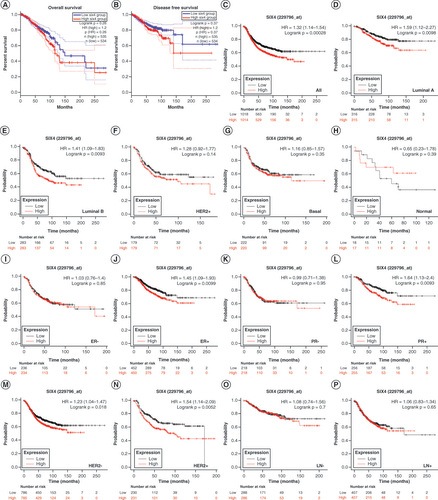
(A) Overall survival. (B) Disease-free survival. (C) Relapse-free survival (RFS) in all patients. (D) Luminal A subtype RFS. (E) Luminal B subtype RFS. (F) HER2+ subtype RFS. (G) Basal-like subtype RFS. (H) Normal-like subtype RFS. (I) ER− patient RFS. (J) ER+ patient RFS. (K) PR− patient RFS. (L) PR+ patient RFS. (M) HER2− patient RFS. (N) HER2+ patient RFS. (O) RFS for patients without lymph node metastasis. (P) RFS for patients with lymph node metastasis.
ER: Estrogen receptor; PR: Progesterone receptor.
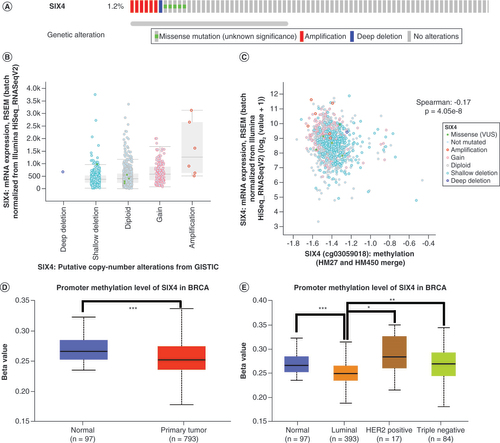
(A) Mutation percentage and types of SIX4 in breast cancer patients from The Cancer Genome Atlas, PanCancer Atlas. (B) Distribution of SIX4 expression in different copy number variation groups. (C) Correlation of SIX4 mRNA expression with copy number variation groups. (D) Promoter methylation level of SIX4 in primary breast cancer compared with normal breast tissue samples. (E) Promoter methylation level of SIX4 in different molecular subtypes of breast cancer.
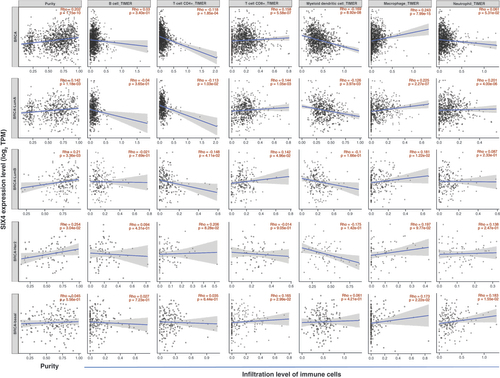
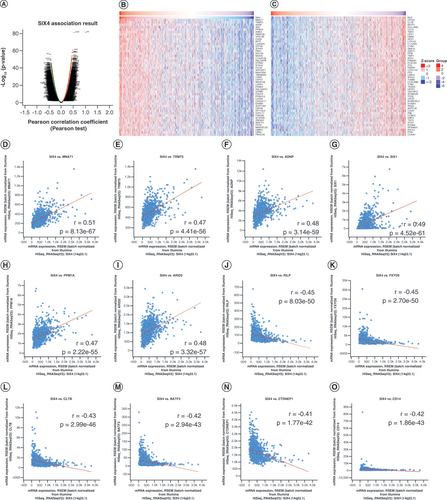
(A) Genes whose expression is correlated with that of SIX4 in breast cancer. (B) Top 50 genes positively correlated with SIX4. (C) Top 50 genes negatively correlated with SIX4. (D)MNAT1. (E)TRMT5. (F)ADNP. (G)SIX1. (H)PPM1A. (I)ARID2. (J)RILP. (K)FXYD5. (L)CLTB. (M)BATF3. (N)CTDNEP1 (DULLARD). (O)CD14.
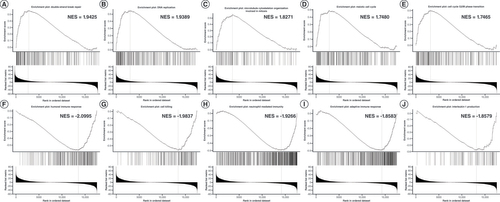
(A) Double-strand break repair. (B) DNA replication. (C) Microtubule cytoskeleton organization involved in mitosis. (D) Meiotic cell cycle. (E) Cell cycle G2/M phase transition. (F) Humoral immune response. (G) Cell killing. (H) Neutrophil-mediated immunity. (I) Adaptive immune response. (J) IL-1 production.
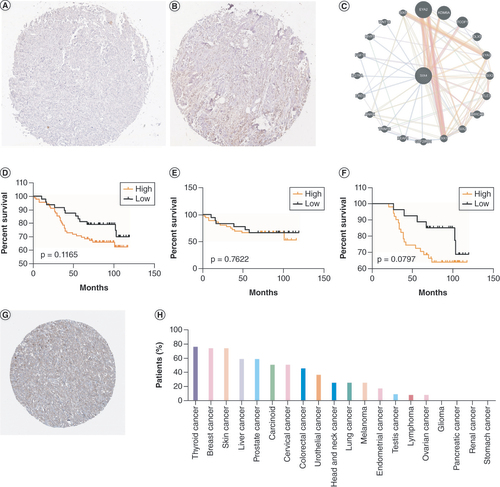
(A & B) Representative staining of SIX4 in breast cancer tissues with (A) low and (B) high expression. (C) Protein–protein interactions of SIX4. (D–F) Overall survival of (D) all breast cancer patients, (E) ER− patients and (F) ER+ patients. (G) Staining of SIX4 in the Human Protein Atlas (HPA031794). (H) The protein expression of SIX4 in different malignancies.
ER: Estrogen receptor.