Figures & data
Table 1.
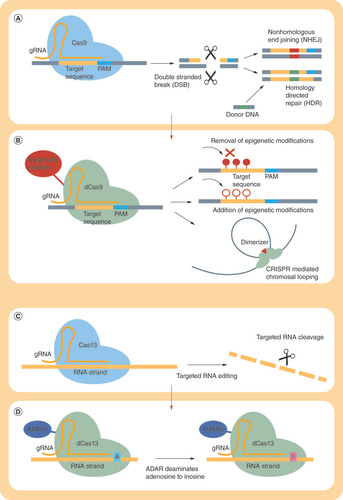
Types of CRISPR systems that have been repurposed for epigenome editing.
Table 2.
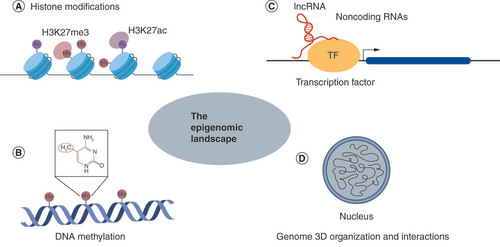
dCas9-mediated epigenome editing involves multiple epigenetic mechanisms.
Table 3.
Applications of CRISPR mediated epigenome editing in cancer research and regenerative medicine.