Figures & data
(A) Molecular mechanisms underlying miRNA-dependent inhibition of gene expression. miRNAs are incorporated in Argonaute proteins, which they guide to mRNAs. Through the interactions of Argonaute proteins with various protein complexes, miRNAs induce translational repression, decapping and deadenylation of the target mRNAs. (B) Transcription factors and epigenetic regulators are preferred targets of miRNAs. We calculated the probability of each gene in a specific functional class being targeted by miRNAs and then plotted the cumulative distribution of the probability values for genes in individual functional categories. Briefly, targeting scores for each human gene and every miRNA seed family were obtained by first averaging the TargetScan aggregate probability of conserved targeting (PCT) scores Citation[33] of all transcripts that are associated with the gene. The probability of a gene to be targeted by at least one miRNA (Pgene) was calculated as: , where PmiR,gene is the probability that the gene is targeted by a specific miRNA miR, calculated as described above. Transcription factors were obtained from the DBD database Citation[157] and epigenetic regulators by extracting genes that were annotated with epigenetic-related gene ontology terms from the AmiGO database Citation[158]. The number of genes in each category is indicated in parentheses. Compared with all genes, epigenetic regulators and transcription factors tend to have higher probabilities of being miRNA targets.
![Figure 1. Mechanisms and preferred targets of miRNAs. (A) Molecular mechanisms underlying miRNA-dependent inhibition of gene expression. miRNAs are incorporated in Argonaute proteins, which they guide to mRNAs. Through the interactions of Argonaute proteins with various protein complexes, miRNAs induce translational repression, decapping and deadenylation of the target mRNAs. (B) Transcription factors and epigenetic regulators are preferred targets of miRNAs. We calculated the probability of each gene in a specific functional class being targeted by miRNAs and then plotted the cumulative distribution of the probability values for genes in individual functional categories. Briefly, targeting scores for each human gene and every miRNA seed family were obtained by first averaging the TargetScan aggregate probability of conserved targeting (PCT) scores Citation[33] of all transcripts that are associated with the gene. The probability of a gene to be targeted by at least one miRNA (Pgene) was calculated as: , where PmiR,gene is the probability that the gene is targeted by a specific miRNA miR, calculated as described above. Transcription factors were obtained from the DBD database Citation[157] and epigenetic regulators by extracting genes that were annotated with epigenetic-related gene ontology terms from the AmiGO database Citation[158]. The number of genes in each category is indicated in parentheses. Compared with all genes, epigenetic regulators and transcription factors tend to have higher probabilities of being miRNA targets.](/cms/asset/4d341365-b9be-448a-b27e-faa8e9e762ee/iepi_a_12324863_f0001.jpg)
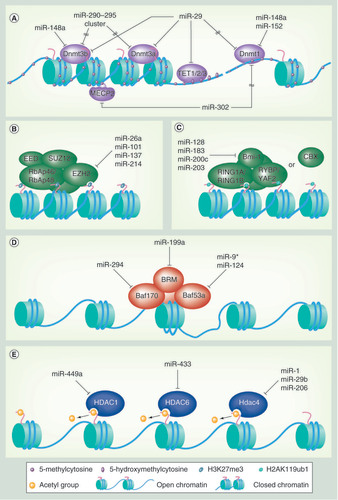
(A) miRNAs that directly or indirectly regulate targets in the DNA methylation/demethylation pathways. (B–E) miRNAs that target a catalytic subunit of (B) PCR2, (C) PCR1, (D) SWI/SNF complex subunits and (E) histone deacetylases.