Figures & data
All figures are viewable in color online at www.tandfonline.com/doi/full/10.2217/epi.13.74
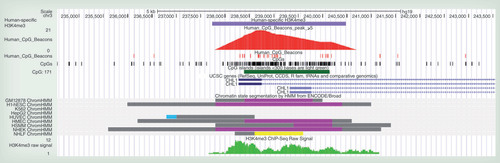
Outer ring: Chromosomes with banding patterns. Dense edge ring: Enredo-Pecan-Ortheus [Citation10,Citation11] h1c1o1 coverage of these chromosomes. Blue peaks: extreme CpG ‘beacon’ peak locations. Human-specific H3K4me3 location with false discovery rate log p values compared to chimpanzee (orange), macaque (green) and overlapping chimpanzee and macaque (olive). Location of ANKRD11, CHL1, DPP10, HAR1A and PSD4 indicated with colocating CpG ‘beacon’ clusters and human-specific H3K4me3. The highlighted yellow wedge is the location of the chromosome 2qFus (2q13–2q14.1) with enrichment of both extreme CpG ‘beacon’ clusters and of Human-specific H3K4me3 peaks (χ2: p = 1.045 × 10-5).
![Figure 2. Human chromosomes 2, 3, 16 and 20.Outer ring: Chromosomes with banding patterns. Dense edge ring: Enredo-Pecan-Ortheus [Citation10,Citation11] h1c1o1 coverage of these chromosomes. Blue peaks: extreme CpG ‘beacon’ peak locations. Human-specific H3K4me3 location with false discovery rate log p values compared to chimpanzee (orange), macaque (green) and overlapping chimpanzee and macaque (olive). Location of ANKRD11, CHL1, DPP10, HAR1A and PSD4 indicated with colocating CpG ‘beacon’ clusters and human-specific H3K4me3. The highlighted yellow wedge is the location of the chromosome 2qFus (2q13–2q14.1) with enrichment of both extreme CpG ‘beacon’ clusters and of Human-specific H3K4me3 peaks (χ2: p = 1.045 × 10-5).](/cms/asset/eef5f4c8-5d84-4f4e-90a0-a07f438f0f03/iepi_a_12324878_f0002.jpg)
This analysis is within the primate-alignable (EPO [Citation10,Citation11]) portion of the human genome. EPO: Enredo-Pecan-Ortheus; FDR: False discovery rate.
![Figure 3. Stronger human-specific H3K4m3 peaks identified (FDR p value) with CpG ‘beacon’ density in both human versus chimpanzee (A), and versus rhesus macaque (B) (Wilcoxon test CpG ‘beacon’ 0 vs > = 20, p = 3.57 × 10-3 chimpanzee and p = 2.50 × 10-2 macaque (p = 0 values capped at 10-200).This analysis is within the primate-alignable (EPO [Citation10,Citation11]) portion of the human genome. EPO: Enredo-Pecan-Ortheus; FDR: False discovery rate.](/cms/asset/1d78347f-4911-4893-a8a6-8b0bfc8e6b19/iepi_a_12324878_f0003.jpg)
CpG ‘beacon’ clusters identify hs-H3K4me3 peaks more successfully than extreme human nucleotide change or studies of selection. Scatter plot of permutated data for random selection of sets of one to 1000 loci from the total of 18,665 human peaks (1000× randomization each) that then overlap the 410 hs-H3K3me4 loci. Upper line: Loess regression line of maximum simulation value; golden points: average for each random selection with average Loess regression line overlaid in grey; red squares: CpG ‘beacon’ clusters; green squares: CpG ‘beacon’ clusters adjusted for the fact that the primate EPO alignment only contains 257 hs-H3K4me3 peaks; blue squares: selective studies – HACNS (HACNS – 0/992) [Citation43] and ANC (ANC – 6/1356) [Citation44], Select_2x, _3x and _4+x are overlaps with studies combined by Akey [Citation42] with loci colocating in 2, 3 or 4 or more studies. HARs are the 49 top HARs [Citation21]. HAR_202 is an expanded set of 202 HARs, with this set the only nonbeacon analysis to perform well, as this larger grouping has a strong overlap with CpG ‘beacons’ (∼85.7% of the hs-H3K4me3 overlapping CpG ‘beacons’ ≥10/kb). ANC: Accelerated noncoding; CB_10: CpG ‘beacon’ clusters ≥10/kb; CB_15: CpG ‘beacon’ clusters ≥15/kb; CB_20: CpG ‘beacon’ clusters ≥20/kb; HAR: Human accelerated regions; HACNS: Human accelerated conserved noncoding; hs-H3K4me3: Human-specific H3K4me3.
![Figure 4. Comparison of studies: proportion of identified loci with overlapping human-specific H3K4me3.CpG ‘beacon’ clusters identify hs-H3K4me3 peaks more successfully than extreme human nucleotide change or studies of selection. Scatter plot of permutated data for random selection of sets of one to 1000 loci from the total of 18,665 human peaks (1000× randomization each) that then overlap the 410 hs-H3K3me4 loci. Upper line: Loess regression line of maximum simulation value; golden points: average for each random selection with average Loess regression line overlaid in grey; red squares: CpG ‘beacon’ clusters; green squares: CpG ‘beacon’ clusters adjusted for the fact that the primate EPO alignment only contains 257 hs-H3K4me3 peaks; blue squares: selective studies – HACNS (HACNS – 0/992) [Citation43] and ANC (ANC – 6/1356) [Citation44], Select_2x, _3x and _4+x are overlaps with studies combined by Akey [Citation42] with loci colocating in 2, 3 or 4 or more studies. HARs are the 49 top HARs [Citation21]. HAR_202 is an expanded set of 202 HARs, with this set the only nonbeacon analysis to perform well, as this larger grouping has a strong overlap with CpG ‘beacons’ (∼85.7% of the hs-H3K4me3 overlapping CpG ‘beacons’ ≥10/kb). ANC: Accelerated noncoding; CB_10: CpG ‘beacon’ clusters ≥10/kb; CB_15: CpG ‘beacon’ clusters ≥15/kb; CB_20: CpG ‘beacon’ clusters ≥20/kb; HAR: Human accelerated regions; HACNS: Human accelerated conserved noncoding; hs-H3K4me3: Human-specific H3K4me3.](/cms/asset/7d2f8eaa-986b-4e3f-aedc-0e604356af53/iepi_a_12324878_f0004.jpg)
The histone methylase PRDM9 directs recombination via motif-specificity, with resultant biased gene conversion within these regions increasing GC content and therefore CpG numbers. Dense CpG regions are able to recruit chromatin-modifying factors such as Cpf1, which lead to the formation of permissive H3K4me3 chromatin domains. Due to the strong association of PRDM9 with recombination [Citation51], ancestral alleles of this rapidly evolving DNA-binding factor [Citation52] are hypothesized to be implicated in this recombination-associated biased gene conversion [Citation39].
![Figure 5. The hypothesis associating potential recombination-driven permissive chromatin formation with CPGi formation via CpG ’beacons’.The histone methylase PRDM9 directs recombination via motif-specificity, with resultant biased gene conversion within these regions increasing GC content and therefore CpG numbers. Dense CpG regions are able to recruit chromatin-modifying factors such as Cpf1, which lead to the formation of permissive H3K4me3 chromatin domains. Due to the strong association of PRDM9 with recombination [Citation51], ancestral alleles of this rapidly evolving DNA-binding factor [Citation52] are hypothesized to be implicated in this recombination-associated biased gene conversion [Citation39].](/cms/asset/ed30dd98-b171-447d-9010-1ec3b7319ae7/iepi_a_12324878_f0005.jpg)
