Figures & data
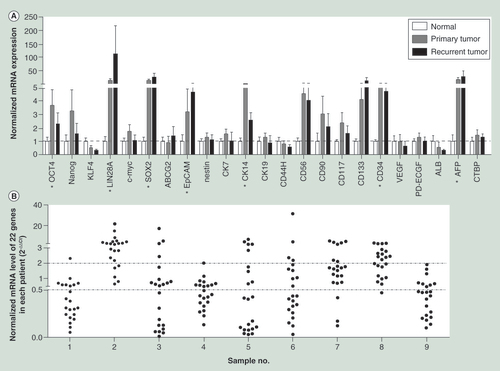
(A) The methylation patterns between the primary and recurrent tumors of Cases 1 and 2, who were HBV negative. (B) The methylation patterns between the primary and recurrent tumors of Cases 3 to 9, who were HBV infection related. (C) The methylation pattern between Hep-11 and Hep-12 cell lines. The Spearman’s rank correlation coefficient test was used to analyze the correlations, p < 0.05 was considered significant.
P: Primary; R: Recurrent.
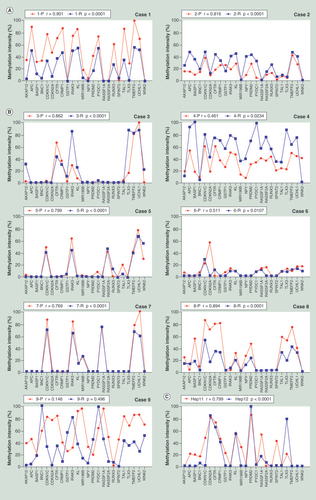
(A) The HBV DNA integration in Case 8. (B) The HBV DNA integration in Case 9. Left panel shows the Alu-PCR results detected by agarose gel electrophoresis separation. Lanes 1–4 represent different PCR results using different Alu-PCR primers. Lane 1: HBV X gene primer as forward and Alu (+) primer as reversed; Lane 2: HBV preC/C gene primer as forward and Alu (+) primer as reversed; Lane 3: HBV X gene primer as forward and Alu (−) primer as reversed; Lane 4: HBV preC/C gene primer as forward and Alu (−) primer as reversed. The right panel shows the sequencing results of the circled bands in the left panel. The arrows indicates the location of HBV–host genome junctions.
P: Primary hepatocellular carcinoma tumor; R: Recurrent hepatocellular carcinoma tumor.
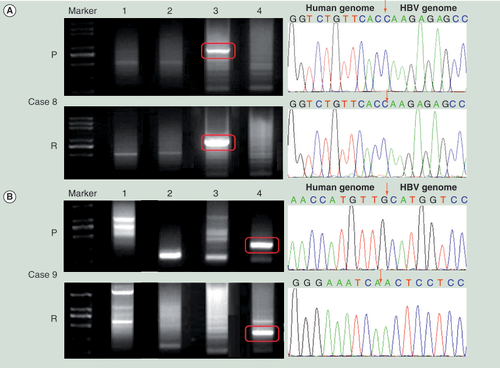
(A) The mRNA expression levels of the 22 cancer stem cell biomarkers in primary and recurrent hepatocellular carcinoma tissues normalized by that in normal liver tissues. The housekeeping gene, CTBP, was also detected as a quality control. (B) The normalized mRNA expression levels of 22 cancer stem cell biomarkers in recurrent tumors compared with that in the primary.
*indicates that the gene’s expression level of either the primary tumor or the recurrent was significantly higher than that of normal liver tissues. The two-tailed unpaired t-test was used to analyze the difference of expression levels in different groups.
p < 0.05 was considered significant.