Figures & data
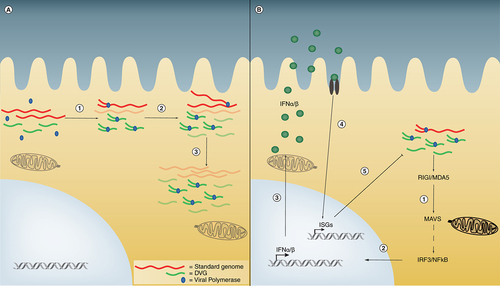
(A) Competition for viral products occurs in cells that contain several copies of standard virus and DVGs. A limited available amount of polymerase and associated proteins randomly binds to viral genomes to begin the replication process (1). Data indicate that viral promoters on DVGs bind the viral polymerase with higher affinity than promoters in standard virus. As DVGs are shorter than standard virus they are also synthesized more rapidly and thus quickly accumulate (2). With a combination of faster transcription and stronger affinity for polymerase DVGs eventually outcompete standard virus to become the predominant species (3), thereby interfering with standard virus replication. (B) In interference by IFN, infected cells first detect DVGs through the RNA sensors RIG-I or MDA5 (1). Signaling through the adaptor protein MAVS leads to IRF3 and NFκB activation and translocation to the nucleus (2). These molecules stimulate the production and secretion of IFNα/β (3) that act in an autocrine or paracrine manner (4) to produce ISGs that inhibit viral replication (5).
DVG: Defective viral genome; IFN: Interferon; ISG: Interferon stimulated gene; MAVS: Mitochondria antiviral-signaling protein.
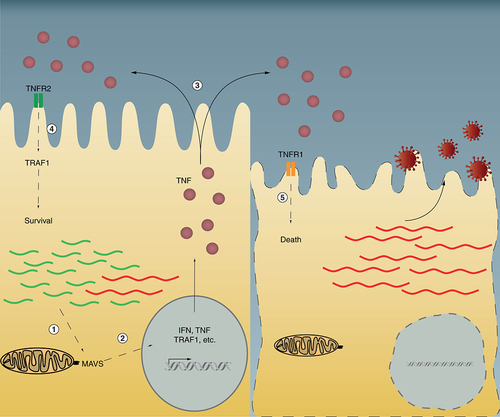
Defective viral genome (DVGs) are sensed by the RIG-I-like receptors RIG-I and MDA5 and activate the downstream signaling component MAVS (1). MAVS signaling leads to the production of TNFR2, TRAF1, IFN, and TNF (2). IFN and TNF are secreted to act in an autocrine or paracrine manner (3). In DVG-high cells expressing TNFR2 and TRAF1, a prosurvival pathway is activated and virus persistence is established. In cells high in standard virus genome but with few or no DVGs the TNFR1 pathway is activated leading to cell death (4).
IFN: Interferon; MAVS: Mitochondria antiviral-signaling protein.
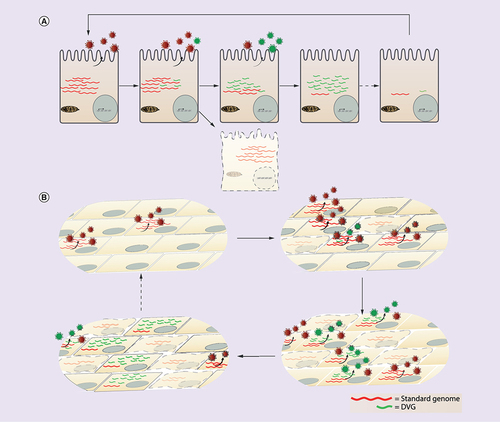
(A) Intracellular cycling begins with a cell infected with standard virus. Upon virus replication DVGs slowly accumulate and are shed in Defective interfering (DI) particles. Cells that accumulate DVGs may escape cell death and instead transition to a DVG-high status where virus is no longer produced and viral proteins are not longer made due to interference. As a result, viral RNA decreases and the cycle is reinitiated given that DVGs are somehow eliminated. (B) In a population model, cycling also begins with a standard virus infection. As the infection propagates, DVGs form and DI particles are shed. Cells failing to produce DVGs die while cells able to replicate DVGs survive accumulating DVGs until reaching a high content accompanied by low standard virus. As DVGs interfere with viral protein production within cells, DVG and virus production diminishes. Cells that have died from infection with standard virus are eventually replaced by new cells allowing the cycle to begin again as standard virus infects those new uninfected cells.
DVG: Defective viral genome.