Figures & data
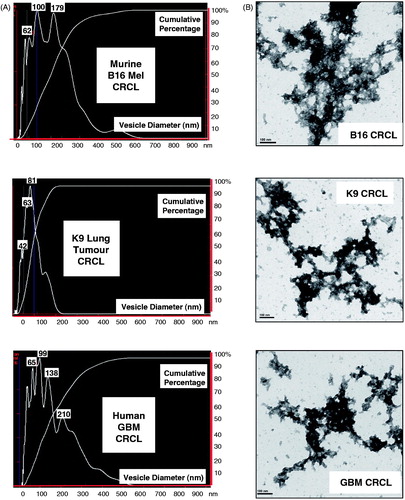
Figure 1. Overlap of proteins found in four human CRCL preparations, and gene ontogeny categorisations of the proteomes. (A) Venn diagram showing the distribution CRCL proteomes amongst each other, ranging from the 95 proteins found in common to all (red circle) to the 143 proteins found only in colorectal cancer (CRC) CRCL, the 76 found in only glioblastoma (GBM) CRCL, the 136 found only in non-small cell lung cancer (NSCLC) CRCL, and the 57 found only in ovarian cancer (OVA) CRCL. Total numbers of proteins identified are also shown. (B) IPA results for ‘top bio functions/diseases and disorders’ are shown for each of the four tumour CRCL proteomes (listed in the lower right of each graph). These categories include ‘cancer, immunological (Immunolog) disease’, ‘neurological disease’, ‘dermatological diseases and conditions (Dermat Dis & Condis)’, ‘haematological (Hemat) disease’, ‘gastrointestinal (GI) disease’, ‘skeletal and muscular (Skel & Musc) disorders’, ‘inflammatory (Inflamm) disease’, and ‘psychological disorders’. X-axis is the −log (p value), with anything higher than 1.25 being statistically significant. (C) IPA results for the 95-protein ‘core’ found in common to all 4 CRCL preparations. Analyses and categories are shown atop each graph. The five categories for ‘networks/associated functions’ include ‘cancer, haematological and immunological (Cancer, Hemat & Immune) disease’, ‘cellular compromise, cellular function and maintenance, post-translational modification (Cell Comp, Funct, Maintenance, PTM)’, ‘cancer, dermatological diseases and conditions, haematological disease (Cancer, Derm Dis & Condits, Hemat Dis)’, ‘immunological disease, inflammatory disease, cellular compromise (Immune/Inflamm Disease, Cell Comp)’, and ‘cardiovascular disease, molecular transport, cell morphology (CV Disease, Mol Transp, Cell Morph)’. The ‘diseases and disorders’ categories overlap with . The ‘canonical pathways’ include ‘remodelling of epithelial adherens junctions (Remod Epith Adh Junct)’, ‘14-3-3-mediated (Sig) signalling’, ‘epithelial adherens junction signalling (Epith Adherens Junct Sig)’, ‘germ cell-Sertoli cell junction signalling (Germ/Sertoli Cell Junct Sig)’, and ‘Sertoli cell-Sertoli cell junction signalling (Sertoli Cell Junct Sig)’. The ‘upstream regulator’ analytic identifies transcriptional regulators associated with the pathways discerned from the proteomic connections. The x-axes are as described in . APP = amyloid beta (A4) precursor protein; MAPT = microtubule-associated protein tau; PSEN1 = presenilin 1; TP53 = tumour protein p53.
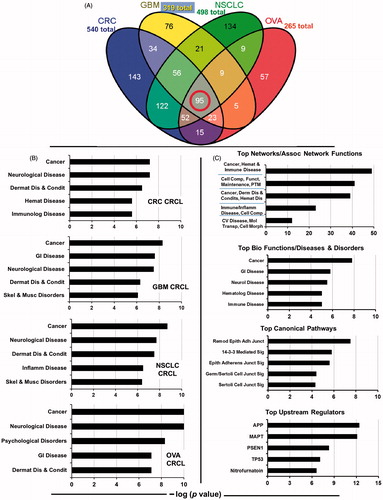
Figure 2. IPA interactomes generated from the 95-protein core catalogued in the top 2 networks/associated functions. Proteins identified in our proteomic search are shown in large red font with gold background. Blue lines indicate known/documented interactions between identified proteins, while light blue lines are interactions between identified proteins and other proteins in the collective category database. Broken lines are indirect interactions. More information is given in the text. ‘Score’ is the −log(p value), and ‘focus molecules’ are initiators of networks. A legend for interpreting the protein ‘shapes’ can be found in Supplementary Figure 1.
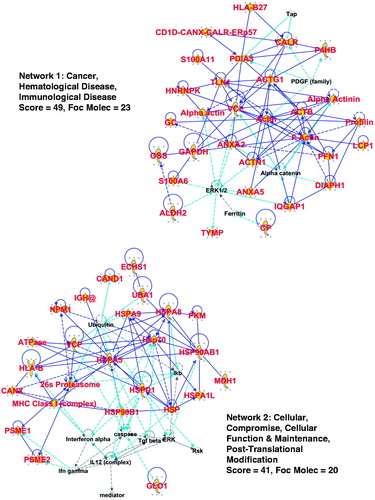
Figure 3. Analyses of CRCL structure by nanoparticle tracking analysis (NanoSight) and by transmission electron microscopy. (A) Particle size distribution profiles are shown for murine (B16 melanoma) CRCL (top), for CRCL prepared from a canine broncho-alveolar adenocarcinoma (middle), and for human glioblastoma CRCL (bottom). Denoted peak heights show vesicle sizes and with the percent of vesicles of that size in that peak (y-axis). (B) TEM of the same CRCL preparations in , depicted in uranyl acetate negative stain; scale bar = 100 nm.