Figures & data
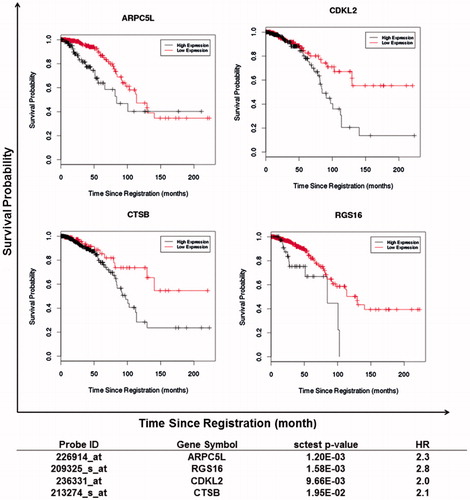
Table 1. Association between gene expression patterns and overall survival in IBC patients.
Table 2. Overview of some of the biological processes associated with differential expression of our genes of interest.
![Figure 1. Wash-in parameter (WiP) maps generated from DCE-MRI in LABC patients. WiP maps are surrogates for perfusion/permeability [Citation7]. The right image shows the CF enhancement pattern which is a uniform enhancement from centre to periphery. The left image is the CP enhancement pattern which is an inhomogeneous ring-type enhancement. The 10–250 scale represents the range of the WiP as previously described in detail [Citation7].](/cms/asset/552e9c22-d4bb-471c-89c0-59ac51b9712c/ihyt_a_1016557_f0001_c.jpg)