Figures & data
FIGURE 1. (a,b) Spectrophotometric scan analysis of plasma circulating nucleic acids and corresponding standards. For the isolation of RNA from plasma samples, the commercial RNA isolation reagent set TRI Reagent BD (T3809) was used. Purified RNAs and related oligonucleotides were employed for spectrophotometric scan analysis. For spectrophotometric scan analysis the solutions of rRNA, DNA polyA, polyC, polyU, polyI:C, polyA:U, and CpG were used. The UV spectra obtained in our study are in a close agreement with recent revised UV extinction coefficients of RNA and related nucleotides.Citation19
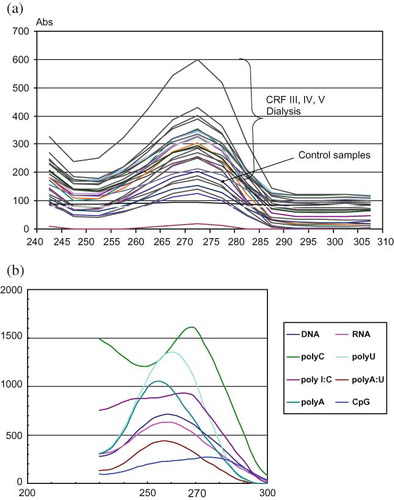
TABLE 1. Main clinical characteristics of investigated groups and circulating RNA level
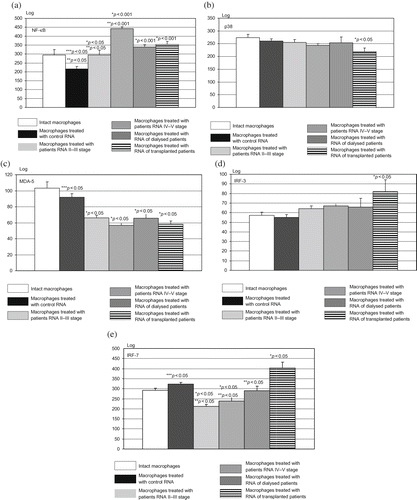
FIGURE 2. (a–e) NF-κB, p38, MDA-5, IRF-3, and IRF-7 level after incubation of residential macrophages with isolated nucleic acids. Residential macrophages, cultured in 24-well plates, were allocated into different groups: the cells exposed to media supplemented with physiological saline solution (intact macrophages); the cells exposed to media supplemented with RNAs purified from plasma of control healthy subjects; the cells exposed to media supplemented with RNAs purified from different groups of patients. The number of samples of each group corresponded to the number of circulating nucleic acid samples, except that untreated group of cells exposed to media and physiological saline solution consisted of 10 samples. The residential macrophages were cultured with isolated nucleic acids for 4 hours, afterwards the cells were transferred into five 96-well plates for determination of NF-κB, p38, MDA-5, IRF-3, and IRF-7. The MFI (logarithmic scale) was determined, and the results presented here are the values of MFI following subtraction of values for unstained matched control cells, analyzed on Victor Perkin Elmer–Wallac multiplate reader. *p < 0.001, statistical significance compared to the cells exposed to media supplemented with RNAs purified from plasma of control healthy subjects. **p < 0.001, statistical significance compared to the cells exposed to media supplemented with RNAs purified from plasma of patients who underwent renal transplantation. ***p < 0.05, statistical significance compared to the cells exposed to media supplemented with physiological saline solution (intact macrophages).