Figures & data
Table I. Sequence of double-stranded oligonucleotides.
Figure 1. Repair of an AP site in a three-lesion cluster with bi-stranded 8-oxoG lesions. (A) Phosphorimaging scan of a polyacrylamide gel showing the repair products from an in vitro BER assay. Band 1; the SSB resulting from incision of the AP site. Band 2; the intermediate band at which the greatest number of nucleotides were incorporated. Band 3; the intact 40 mer band, either before incision of the AP site or following rejoining. Due to a loading error sample AP.8-oxoG-2/8-oxoG + 1 60 min is located in lane ** and sample AP.8-oxoG-2/8-oxoG-3 60 min is located in lane *. (B) and (C) Graphical representation of the rejoined band following repair of the AP site, over time. The horizontal line in C is to show the base line from which the repair of the clusters AP.8-oxoG-2/8-oxoG-1 and AP.8-oxoG-2/8-oxoG-3 were measured. The error bars represent standard deviations of three independent experiments.
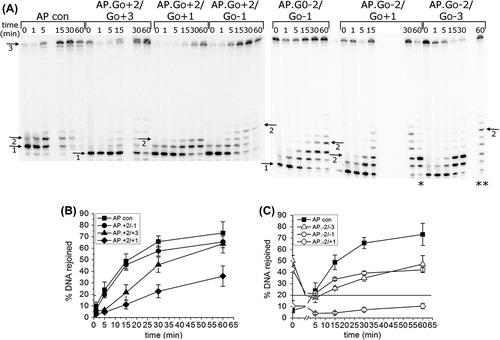
Figure 2. Activity of polymerase β following incision of a control AP site and incision of an AP site in the AP.8-oxoG + 2/8-oxoG + 3 cluster. Polymerase β activity was measured by incorporation of nucleotides following incision of the AP site. The error bars represent standard deviations of three independent experiments.
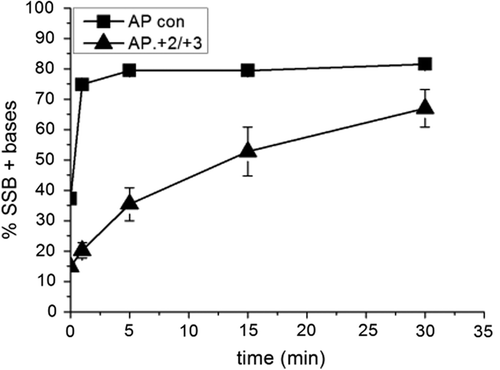
Figure 3. Intermediates of repair of an AP site in a three-lesion cluster with bi-stranded 8-oxoG lesions. (A) and (B) Graphical representation of the SSB + 1 intermediate, over time. (C), (D) and (E) Graphical representation of the intermediate with the greatest number of incorporated nucleotides for each of the three-lesion clusters, over time. The error bars represent standard deviations of three independent experiments.
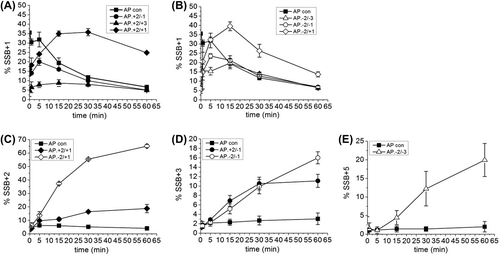
Figure 4. Repair of a three-lesion cluster with an AP site and bi-stranded 8-oxoG lesions. (A) Phosphorimaging scan of a native polyacrylamide gel showing the repair products from an in vitro BER assay with CHO-K1 nuclear extracts. Band 1; double-stranded 40 mer oligonucleotide. Band 2; expected site of shortened double-stranded oligonucleotide resulting from a DSB, if one was formed. (B) Phosphorimage scan of a native polyacrylamide gel showing the repair products from an in vitro BER assay reconstituted with purified BER proteins, in the absence (−) and presence (+) of OGG1 The repair assay was performed for 30 min. Band 1; double-stranded 40 mer oligonucleotide. Band 2; expected site of shortened double-stranded oligonucleotide resulting from a DSB, if one had been formed.
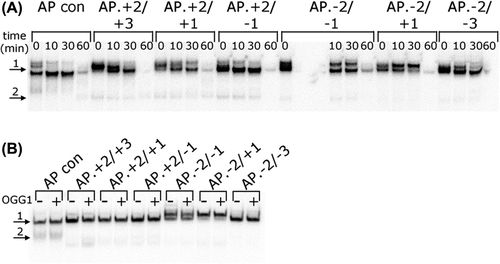
Figure 5. SP repair of an AP site in a three-lesion cluster with bi-stranded 8-oxoG lesions. Deoxynucleotides were substituted with di-deoxynucleotides such that only the SSB + 1 nucleotide could be incorporated, inhibiting LP repair. (A) and (B) Graphical representation of the rejoined band following repair of the AP site, over time. (C) and (D) Graphical representation of the SSB + 1 intermediate, over time. The error bars represent standard deviations of three independent experiments.
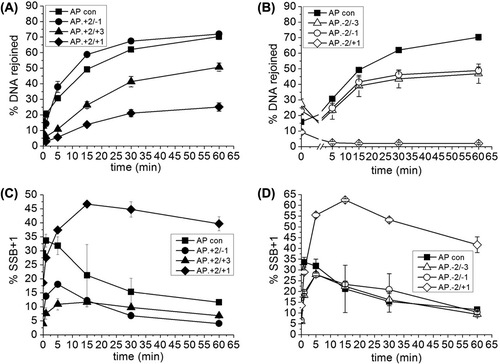
Figure 6. Mutation frequency of a single uracil residue, a single 8-oxoG lesion and a three-lesion cluster of an AP site with bi-stranded 8-oxoG lesions in MutY/Fpg null E. coli. The mutation frequency was calculated by dividing the number of individual mutations found in the oligonucleotide sequence ligated in to pUC19 plasmid by the total number of oligonucleotides sequences sequenced. Some plasmids contained two oligonucleotide sequences. (A) The total number of mutations. The error bars represent standard deviations of three independent experiments. (B) Mutations at either of the 8-oxoG lesions or the uracil residue. (C) Mutations other than those at the 8-oxoG lesions or the uracil residue.
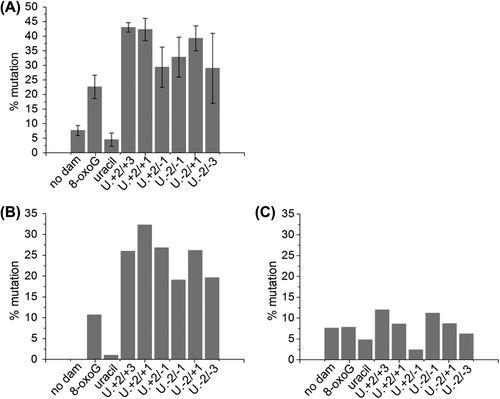
Table II. Mutations detected in oligonucleotides ligated into plasmids, recovered from E. coli.
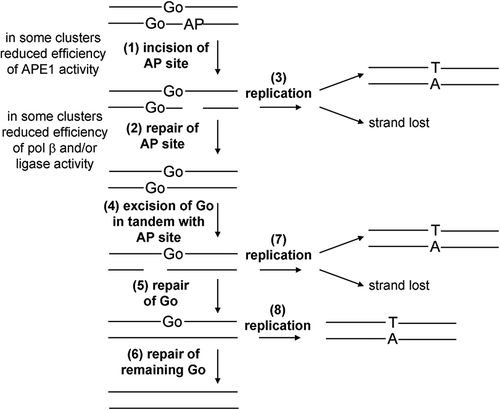
Figure 7. Schematic representation of the repair of the three-lesion cluster in E. coli. Initially the AP site is cleaved (1) and then repaired (2). If replication (3) occurs before repair of the AP site then there is the potential for the SSB.8-oxoG containing strand to be lost or for mutations to arise from the single 8-oxoG containing strand (3). Following repair of the AP site the 8-oxoG lesions are repaired sequentially (4–6). Again if repair is not completed by replication (7 and 8) there is potential for strand loss or mutation induction.