Figures & data
Figure 1. (a) Sequence alignment between the turkey β2AR (adrb_melga) and human βARs (adrb1/2/3_human). The N- and C-termini of the sequences were removed in the alignment, because there is no significant homology between them. Amino acid residues coloured red in the turkey sequence correspond with mutations present in tβ2AR-m23 (R68S, M90V, Y227A, A282L, F327A and F338M), whereas those coloured in blue correspond with other thermostabilizing mutations. Letters above these residues correspond to the position of the residues in (b). Pink rectangles below the sequence alignment indicate the positions of transmembrane domains and the green rectangle corresponds with the position of helix 8. (b) Structure of the turkey β2AR (PDB accession number 2VT4) with the side chains of thermostabilizing mutations represented as space filling models. Side chains in grey are thermostabilizing when mutated to alanine. The five magenta side chains correspond to the five thermostabilizing mutations found in tβ2AR-m23, i.e., R68S, M90V, Y227A, F327A and F338M; the mutation A282L is not shown because this region of the receptor was disordered in this crystal form. The N- and C-termini are labelled, along with the transmembrane α-helices. The model is in rainbow coloration with the N terminus in blue and the C terminus in red. The black sphere is a Na+ ion. This Figure is reproduced in colour in Molecular Membrane Biology online.
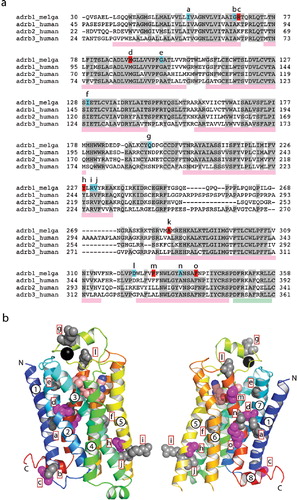
Figure 2. Expression and binding assays of turkey β2AR and human βARs expressed in HEK293 cells. The number of receptors per cell was calculated from single-point ligand binding assays using [3H]DHA at a final concentration of 80 nM performed either on membranes (a) or on cells solubilized with 2% DDM (b). Cells were transfected either with empty vector (V), turkey β2AR (tβ2), human β2AR1-463 (hβ2) or human β2AR (hβ2). Experiments were performed in duplicate and the error bars represent the standard error of the mean (SEM). This Figure is reproduced in colour in Molecular Membrane Biology online.
![Figure 2. Expression and binding assays of turkey β2AR and human βARs expressed in HEK293 cells. The number of receptors per cell was calculated from single-point ligand binding assays using [3H]DHA at a final concentration of 80 nM performed either on membranes (a) or on cells solubilized with 2% DDM (b). Cells were transfected either with empty vector (V), turkey β2AR (tβ2), human β2AR1-463 (hβ2) or human β2AR (hβ2). Experiments were performed in duplicate and the error bars represent the standard error of the mean (SEM). This Figure is reproduced in colour in Molecular Membrane Biology online.](/cms/asset/90384ae9-b1f9-420d-aca0-337e81cb1e36/imbc_a_420996_f0002_b.jpg)
Figure 3. The effect of increasing detergent concentrations on the [3H]DHA binding activity of βARs. Receptors were transiently expressed in HEK293 cells, which were subsequently solubilized at 4°C in different concentrations of DDM (0.01%, 0.05%, 0.1%, 0.5% and 1%); the amount of solubilized functional receptor was determined from a single-point binding assay performed at 4°C using 80 nM [3H]DHA; (a) turkey β2AR, (b) human β2AR1-463 and (c) human β2AR. Experiments were performed in duplicate and the error bars represent the SEM.
![Figure 3. The effect of increasing detergent concentrations on the [3H]DHA binding activity of βARs. Receptors were transiently expressed in HEK293 cells, which were subsequently solubilized at 4°C in different concentrations of DDM (0.01%, 0.05%, 0.1%, 0.5% and 1%); the amount of solubilized functional receptor was determined from a single-point binding assay performed at 4°C using 80 nM [3H]DHA; (a) turkey β2AR, (b) human β2AR1-463 and (c) human β2AR. Experiments were performed in duplicate and the error bars represent the SEM.](/cms/asset/baf73ffb-ddcd-4061-b93b-ef68ef88a107/imbc_a_420996_f0003_b.jpg)
Figure 4. The stability of βARs with and without thermostabilizing mutations. Receptors were solubilized from transiently transfected cells using either 0.1% (red squares) or 0.01% DDM (blue triangles). Samples were then heated at the specified temperature for 30 minutes, quenched on ice and the amount of receptor remaining determined by a single-point ligand binding assay using 80 nM [3H]DHA performed in duplicate; (a) turkey β2AR, (b) human β2AR1-463, (c) human β2AR, (d) turkey β2AR-m23, (e) human β2AR1-463-m23, (f) human β2AR-m23. Stability curves were analyzed by non-linear regression using Prism (GraphPad) to determine the Tms (). This Figure is reproduced in colour in Molecular Membrane Biology online.
![Figure 4. The stability of βARs with and without thermostabilizing mutations. Receptors were solubilized from transiently transfected cells using either 0.1% (red squares) or 0.01% DDM (blue triangles). Samples were then heated at the specified temperature for 30 minutes, quenched on ice and the amount of receptor remaining determined by a single-point ligand binding assay using 80 nM [3H]DHA performed in duplicate; (a) turkey β2AR, (b) human β2AR1-463, (c) human β2AR, (d) turkey β2AR-m23, (e) human β2AR1-463-m23, (f) human β2AR-m23. Stability curves were analyzed by non-linear regression using Prism (GraphPad) to determine the Tms (Table I). This Figure is reproduced in colour in Molecular Membrane Biology online.](/cms/asset/3a2307c2-f667-442b-b04f-1b341c849d8a/imbc_a_420996_f0004_b.jpg)
Table I. Apparent Tms of β receptors solubilized in different concentrations of dodecylmaltoside (DDM) and decylmaltoside (DM).
Table II. Affinity of agonist and antagonist binding to detergent solubilized βARs.
Figure 5. Resistance of the human β2AR1-463 and β2AR to decylmaltoside. (a) Receptors were solubilized with different amounts of decylmaltoside (DM) and the ligand binding activity measured using 80 nM [3H]DHA. Red bars, hβ2AR1-463; pink bars, hβ2AR1-463-m23; dark blue bars, hβ2AR and light blue bars, hβ2AR-m23. The DM concentrations used are as follows: a, 0.1%; b, 0.3%; c, 1%. (b) The stability of the hβ2AR1-463 and hβ2AR solubilized in decylmaltoside (DM) was determined by incubating the sample for 30 minutes at different temperatures, quenching on ice and determining the amount of remaining receptors by assaying with 80 nM [3H]DHA: red, hβ2AR1-463; pink, hβ2AR1-463-m23; dark blue, hβ2AR and light blue, hβ2AR-m23. Stability curves were analysed by non-linear regression using Prism (GraphPad) to determine the Tms (). This Figure is reproduced in colour in Molecular Membrane Biology online.
![Figure 5. Resistance of the human β2AR1-463 and β2AR to decylmaltoside. (a) Receptors were solubilized with different amounts of decylmaltoside (DM) and the ligand binding activity measured using 80 nM [3H]DHA. Red bars, hβ2AR1-463; pink bars, hβ2AR1-463-m23; dark blue bars, hβ2AR and light blue bars, hβ2AR-m23. The DM concentrations used are as follows: a, 0.1%; b, 0.3%; c, 1%. (b) The stability of the hβ2AR1-463 and hβ2AR solubilized in decylmaltoside (DM) was determined by incubating the sample for 30 minutes at different temperatures, quenching on ice and determining the amount of remaining receptors by assaying with 80 nM [3H]DHA: red, hβ2AR1-463; pink, hβ2AR1-463-m23; dark blue, hβ2AR and light blue, hβ2AR-m23. Stability curves were analysed by non-linear regression using Prism (GraphPad) to determine the Tms (Table I). This Figure is reproduced in colour in Molecular Membrane Biology online.](/cms/asset/775c28c5-b7d8-4f86-af8a-f6295884399a/imbc_a_420996_f0005_b.jpg)
Figure 6. Competition binding assays of human βARs with and without the m23 thermostabilizing mutations. Increasing amounts of the agonists, isoprenaline (a) and adrenaline (b), and the antagonist cyanopindolol (c), were used to compete with binding of [3H]DHA present at a concentration three times greater than the KD for each receptor (Supplementary Table S3, online version only): red squares, hβ2AR1-463; pink circles, hβ2AR1-463-m23; dark blue triangles, hβ2AR; pale blue inverted triangles, hβ2AR-m23. Data were analyzed with GraphPad Prism and the Kis determined (). This Figure is reproduced in colour in Molecular Membrane Biology online.
![Figure 6. Competition binding assays of human βARs with and without the m23 thermostabilizing mutations. Increasing amounts of the agonists, isoprenaline (a) and adrenaline (b), and the antagonist cyanopindolol (c), were used to compete with binding of [3H]DHA present at a concentration three times greater than the KD for each receptor (Supplementary Table S3, online version only): red squares, hβ2AR1-463; pink circles, hβ2AR1-463-m23; dark blue triangles, hβ2AR; pale blue inverted triangles, hβ2AR-m23. Data were analyzed with GraphPad Prism and the Kis determined (Table II). This Figure is reproduced in colour in Molecular Membrane Biology online.](/cms/asset/191388c5-187f-40e2-80ce-35ff76313956/imbc_a_420996_f0006_b.jpg)