Figures & data
Table I. Enzyme activity of BicA dual reporter fusions.
Table II. Predicted topologies for BicA using available prediction programs.
Figure 1. The topology map for the Synechococcus PCC7002 Na+-dependent HCO3- transporter, BicA. Experimentally determined phoA/lacZ mapping positions () are shown as black (cytoplasmic), mid-grey (TMH located) or light grey (periplasmic); in the online version these are shown as blue, purple and red, respectively. The inset shows a WebLogo (weblogo.berkeley.edu) representation of the consensus of the ungapped alignment of the 50 residue loop between TMH 8 and 9; all the SulP/SLC26A members used to generate the phylogenetic tree shown in were used and BicA numbering is shown. This region corresponds to the second signature sequence identified by Saier et al. [Citation23] for the SulP family; the most highly conserved subset, NSNKELIGQGLGN (279-291), is highlighted. This Figure is reproduced in colour in the online version of Molecular Membrane Biology.
![Figure 1. The topology map for the Synechococcus PCC7002 Na+-dependent HCO3- transporter, BicA. Experimentally determined phoA/lacZ mapping positions (Table I) are shown as black (cytoplasmic), mid-grey (TMH located) or light grey (periplasmic); in the online version these are shown as blue, purple and red, respectively. The inset shows a WebLogo (weblogo.berkeley.edu) representation of the consensus of the ungapped alignment of the 50 residue loop between TMH 8 and 9; all the SulP/SLC26A members used to generate the phylogenetic tree shown in Figure 3 were used and BicA numbering is shown. This region corresponds to the second signature sequence identified by Saier et al. [Citation23] for the SulP family; the most highly conserved subset, NSNKELIGQGLGN (279-291), is highlighted. This Figure is reproduced in colour in the online version of Molecular Membrane Biology.](/cms/asset/5131875b-2944-4eaf-b3a6-ac676594d9bb/imbc_a_440190_f0001_b.jpg)
Figure 2. A simplified version of the topology map for BicA (Synechococcus PCC7002) showing the positions of positive charged residues and three highly conserved residues that are discussed in the text, E283 and N291, that are implicated in human disease and T489, which is a putative phosphorylation site.
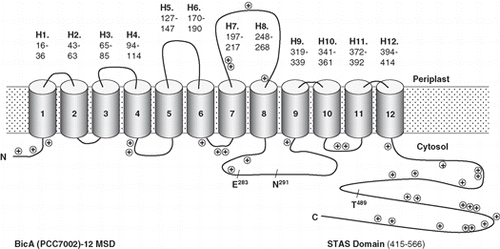
Figure 3. Phylogenetic tree showing the relationship of the cyanobacterial BicA family (30 members) to the Arabidopsis SulP sulphate transporter family (13 members), the human SLC26A family (9 members) and a selection of bacterial BicA-like proteins (13 members). Protein sequences were aligned using ClustalW and nearest neighbour-joining routines in MEGA4 software (www.megasoftware.net). The scale marker represents 0.2 substitutions per residue.
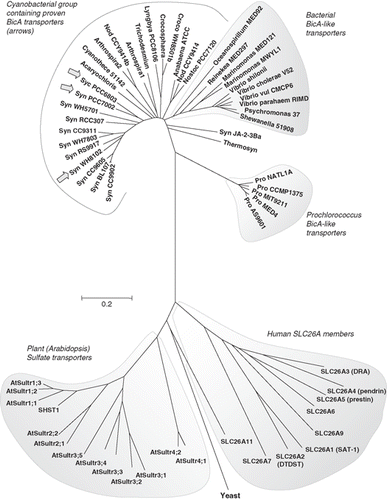
Figure 4. BicA (Synechococcus PCC7002) aligned to an extensively investigated member of the plant sulphate transporter family (SHST1, Stylosanthes hamata) and a member of the human SLC26A family (DTDST, SLC26A2) that is linked to the disease, diastrophic dysplasia. SCAMPI-msa predicted TMH regions are shaded grey based on separate queries for each protein. The two disease-causing residues, E283 and N291, and the putative phosphorylation site T489, are shown.
