Figures & data
Table I. Dual reporter activity for fusions within GtrIc.
Figure 1. DAS predictions for potential presence of membrane spanning segments within GtrIc. Twelve transmembrane α-helices were detected at the loose cut-off (dotted red line). Peak signals are annotated according to these measurements. At the strict cut-off (solid blue line), 13 transmembrane α-helices were identified. DAS is used to represent the consensus output, with consistent plots also observed in HMMTOP, PHDhtm, TMHMM, and TMpred. This Figure is reproduced in colour in the online version of Molecular Membrane Biology.
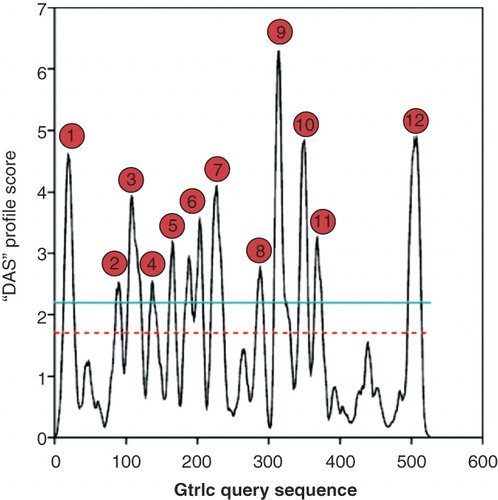
Figure 2. Determination of the GtrIc membrane topology. (a) Various strategies were used to construct pho-lac fusions at different codons of gtrIc. Replacement of the C-terminus of full length gtrIc by PCR (top); random truncation fusions obtained by nested deletion (middle); pho-lac inserted in-frame into gtrIc sequence as a sandwich fusion (bottom). (b) Experimentally verified topology model of GtrIc resolved through computerised modelling and corrected by scanning with pho-lac fusions. Truncation gtrIc/pho-lac fusions (spots) and sandwich gtrIc/pho-lac/gtrIc fusions (encapsulated spots) are coloured according to DI plate observations with their respective residue position. The location of D/ExD/E and KK motifs copies are also provided (green boxes). A larger periplasmic loop 2 was detected relative to the consensus model, along with a double membrane dipping loop centred between the N- and C- termini and ten transmembrane α-helices. This Figure is reproduced in colour in the online version of Molecular Membrane Biology.
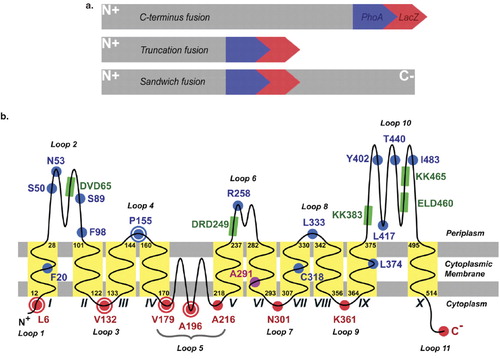
Figure 3. Alignment of Gtr[type] periplasmic sequences. The topologies of GtrII and GtrV were experimentally elucidated in previous studies [Citation14,Citation15]. Identification of loop 2 and large C-terminal loop sequences in GtrI, GtrIV and GtrX was based on preliminary analyses by Lehane et al. [Citation15] by using predictive software and sequence homologies between GtrI and GtrX with GtrII and GtrV, respectively. (a) The large N-terminal loop 2 of all known Gtr[type] members shows a conserved D/ExD/E motif. (b) Conserved KK (dilysine) motifs are seen in the large C-terminal loops of all Gtr[type] proteins except for GtrIV. Two KK motif copies are detected in the same loop of GtrI, GtrIc and GtrV. This Figure is reproduced in colour in the online version of Molecular Membrane Biology.
![Figure 3. Alignment of Gtr[type] periplasmic sequences. The topologies of GtrII and GtrV were experimentally elucidated in previous studies [Citation14,Citation15]. Identification of loop 2 and large C-terminal loop sequences in GtrI, GtrIV and GtrX was based on preliminary analyses by Lehane et al. [Citation15] by using predictive software and sequence homologies between GtrI and GtrX with GtrII and GtrV, respectively. (a) The large N-terminal loop 2 of all known Gtr[type] members shows a conserved D/ExD/E motif. (b) Conserved KK (dilysine) motifs are seen in the large C-terminal loops of all Gtr[type] proteins except for GtrIV. Two KK motif copies are detected in the same loop of GtrI, GtrIc and GtrV. This Figure is reproduced in colour in the online version of Molecular Membrane Biology.](/cms/asset/9f2bf423-fd39-4dc1-b203-f0fcf686efa1/imbc_a_455689_f0003_b.jpg)
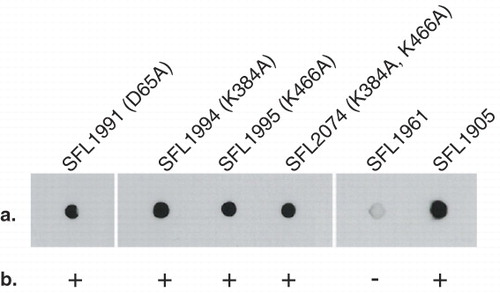
Figure 4. Functional analysis of GtrIc mutants. Transformed SFL1416 serotype 1a harbouring mutated GtrIc constructs were assayed for host 1a to 1c serotype conversion. GtrIc point mutations are annotated in parentheses with their SFL1416-derived host. Comparisons of mutant strains can be made with the SFL1961 containing empty pBCSK+ vector and with SFL1905 containing wild-type gtrIc in pNV1650. (a) Colony immunoblots showing detection of S. flexneri serotype 1c LPS in all mutant strains. (b) Slide agglutinations complemented colony immunoblot readings, showing GtrIc activity in all mutants. Binary scoring was used for positive and negative agglutination tests.