Figures & data
Table I. Primers used for generating SbtA (6803) phoA/lacZ constructs.
Table II. Enzyme activities of SbtA dual reporter fusions.
Figure 1. Topology map of the Na+-dependent HCO3 - transporter, SbtA, from Synechocystis PCC6803. Experimentally determined phoA/lacZ mapping positions are shown as black (internal), mid-grey (TMH located) or light grey (external) based on the scored locations in (primarily based on AP/BG enzyme assay ratios); in the online version these are shown as blue, purple and red, respectively. The positions of positive charges on the inter-helical loops are also shown. This Figure is reproduced in colour in the online version of Molecular Membrane Biology. In addition, a Supplementary residue-based Figure (Supplementary Figure 1) is also provided online.
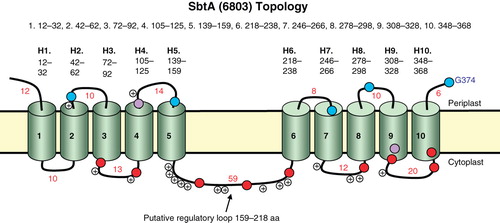
Figure 2. The locations of the ten TMHs in the SbtA from Synechocystis PCC6803 based on experimentally determined phoA/lacZ mapping positions and the SCAMPI-msa prediction model. ‘i’ refers to cytoplasmic domains, while ‘o’ refers to periplasmic domains and grey blocks denote TMH domains.

Figure 3. Alignment of the two halves of the 5 + 5 repeated structure of SbtA-6803. The region encompassing helices 1–5 aligned to the region encompassing helices 6–10 of SbtA.
Figure 4. Simple cartoon showing the inversion of each helix in the repeat (H6–10) relative to each helix in the H1–5 region.
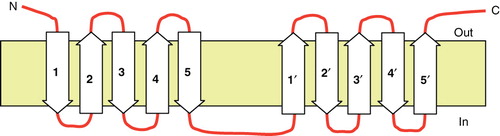
Figure 5. Phylogenetic tree showing the relationship of the extended SbtA family (95 prokaryotic members shown). Protein sequences were aligned using ClustalW and organized into a phylogenetic tree using nearest neighbour-joining routines in MEGA4 software (www.megasoftware.net). The scale marker represents 0.05 substitutions per residue. A list of defined abbreviations is available in Supplementary Table I, available online. This Figure is reproduced in colour in the online version of Molecular Membrane Biology.
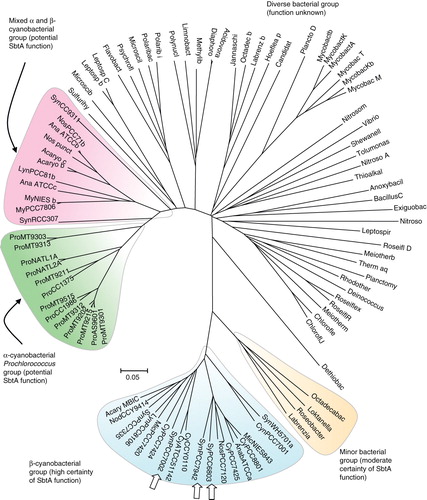
Figure 6. Alpha-helical wheel projections of the ten TMH helices from SbtA-6803. The N-terminal residue in each helix is located at 12 o'clock position each following residue precedes 100° clockwise. A central arrowhead indicates the face with the highest hydrophobic moment (HM); charge residues are highlighted with a star symbol. Hydrophilic (circles), hydrophobic (diamonds), negatively charged (triangles) and positively charged (pentagons) residues are shown. This Figure is reproduced in colour in the online version of Molecular Membrane Biology.
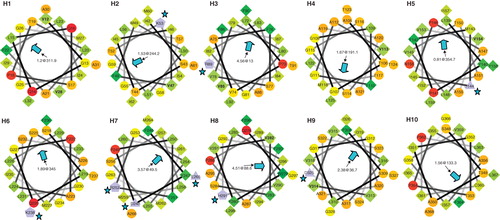
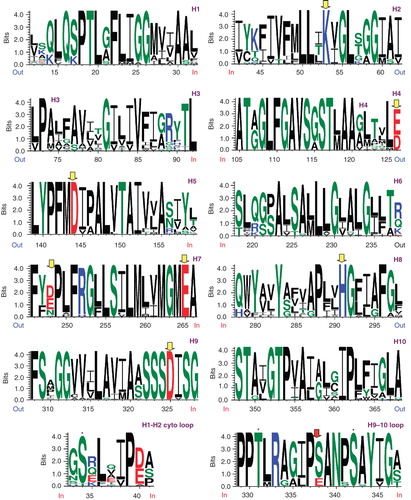
Figure 7. Weblogo representations of aligned helices 1–10 from the 22 member set of high probability SbtA homologs () and including the cytoplasmic loops H1–H2 and H9–H10 in the bottom panel. Conserved charge residues are highlighted with grey (yellow) arrowheads. Conserved S/T residues are indicated with asterisks (bottom panel). This Figure is reproduced in colour in the online version of Molecular Membrane Biology.