Figures & data
Figure 1. Numbers of published polytopic integral membrane protein structures determined by NMR spectroscopy up to the end of the year 2010.
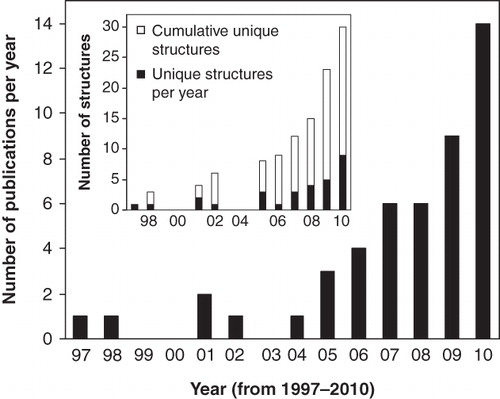
Table I. Details for the structures of polytopic integral membrane proteins determined by NMR spectroscopy published up to the end of the year 2010: Protein information, NMR conditions, PDB entry number.
Table II. Details for the structures of polytopic integral membrane proteins determined by NMR spectroscopy published up to the end of the year 2010: NMR structural restraints. The numbering of the entries in this table corresponds with those in .
Figure 2. NMR structures of β-barrel membrane proteins. Structures of (A) OmpX in DHPC, (B) OmpA from E. coli in DPC, (C) OmpA from Klebsiella pneumoniae in DHPC (D) PagP in β-OG, (E) OmpG in DPC and (F) VDAC1 in LDAO. All pictures were produced using the PDB file and PDB Protein Workshop 3.9 (Moreland et al. Citation2005). This Figure is reproduced in colour in Molecular Membrane Biology online.
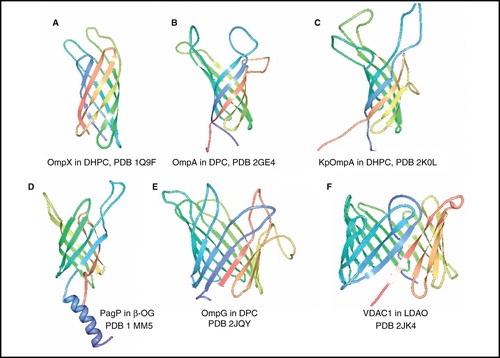
Figure 3. NMR structures of α-helical membrane proteins with two identical α-helices (homodimers). Structures of Glycophorin A in DPC, ζζ TM domain (28–60) in SDS, BNip3 TM domain (154–188) in DPC/DPPC, ErbB2 TM domain (641–684) in DMPC/DHPC, ErbB1/ErbB2 TM domain (634–677/641–684) in DMPC/DHPC, EphA1 TM domain (536–573) in DMPC/DHPC, EphA2 TM domain (523–563) in DMPC/DHPC. All pictures were produced using the PDB file and PDB Protein Workshop 3.9 (Moreland et al. Citation2005). This Figure is reproduced in colour in Molecular Membrane Biology online.
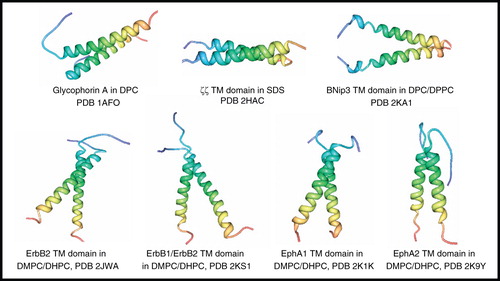
Figure 4. NMR structures of M2 proton channel from influenza A virus. Structures of (A) residues 26–43 + amantadine in DMPC/DMPG by solid-state NMR, (B) residues 18–60 + rimantadine in DHPC by solution-state NMR showing four molecules of bound rimantadine, (C) residues 18–60, S31N + rimantadine in DHPC by solution-state NMR, (D) residues 18–60, V27A by solution-state NMR, (E) residues 22–46 + amantadine in DLPC by solid-state NMR showing a single molecule of amantadine in its high-affinity site and (F) residues 22–62 in DOPC/DOPE by solid-state NMR. Pictures A–D and F were produced using the PDB file and PDB Protein Workshop 3.9 (Moreland et al. Citation2005). Picture E is adapted by permission from Macmillan Publishers Ltd: Nature (Cady et al. Citation2010; copyright 2010). This Figure is reproduced in colour in Molecular Membrane Biology online.
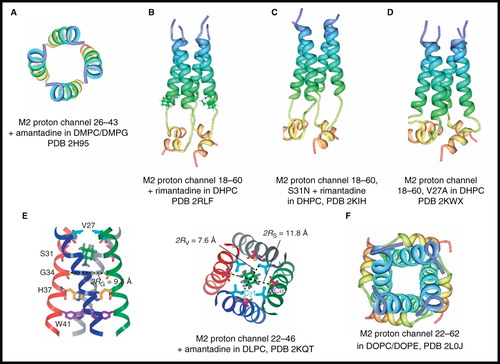
Figure 5. NMR structure of BM2 proton channel from influenza B virus. Structure of BM2 transmembrane domain (residues 1–33) in DHPC; (A) view from the membrane plane showing the occluding residues Phe5 and Trp23 and (B) view from the cytosol. Structure of BM2 cytoplasmic domain (residues 26–109) in LMPG (C). These pictures were produced using the PDB files and PDB Protein Workshop 3.9 (Moreland et al. Citation2005). This Figure is reproduced in colour in Molecular Membrane Biology online.
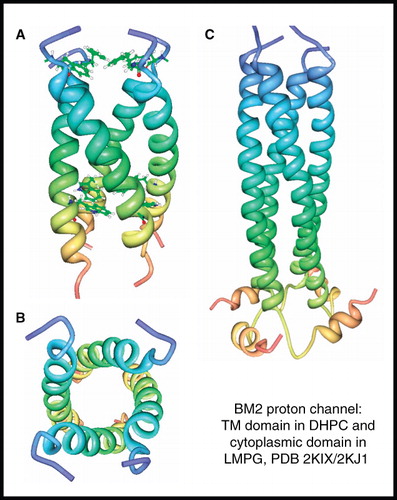
Figure 6. NMR structure of phospholamban homopentamer. Structure of phospholamban homopentamer in DPC; (A) view from the membrane plane and (B) view from the cytosol. These pictures were produced using the PDB file and PDB Protein Workshop 3.9 (Moreland et al. Citation2005). This Figure is reproduced in colour in Molecular Membrane Biology online.
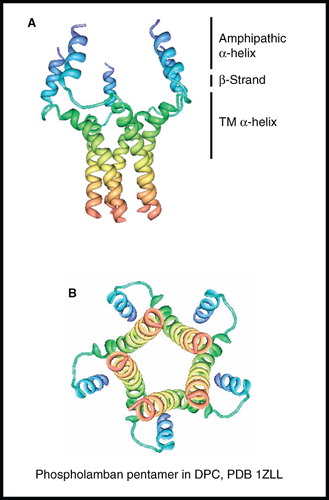
Figure 7. NMR structures of α-helical membrane proteins with two unique α-helices. Structures of (A) subunit c of F1F0 ATP synthase in CHCl3/MeOH/H2O, (B) MerF in SDS by solution-state NMR, (C) MerF in 14-O-PC/6-O-PC by solid-state NMR, (D) Ste2P (31–110) TM1-2 of yeast α-factor receptor in LPPG, (E) integrin αIIbβ3 complex in bicelles, (F) integrin αIIbβ3 complex in CD3CN/H2O, (G) ArcB TM domain (1–115) in LMPG, (H) QseC TM domain (1–185) in LMPG. All pictures were produced using the PDB file and PDB Protein Workshop 3.9 (Moreland et al. Citation2005). This Figure is reproduced in colour in Molecular Membrane Biology online.
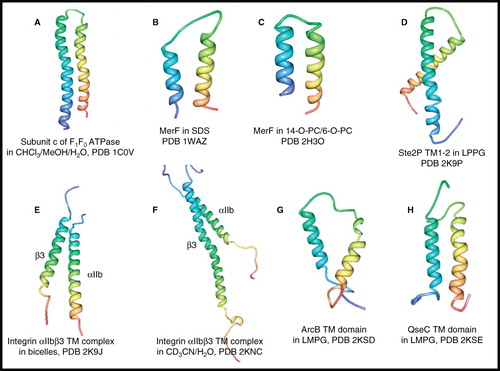
Figure 8. NMR structure of potassium channel KcsA. Structure of residues 1–132 in complex with charybdotoxin in DPC; (A) view from the membrane plane also showing residues involved in channel gating, including the pH sensor His25 and (B) view from the cytosol. These pictures were produced using the PDB file and PDB Protein Workshop 3.9 (Moreland et al. Citation2005). This Figure is reproduced in colour in Molecular Membrane Biology online.
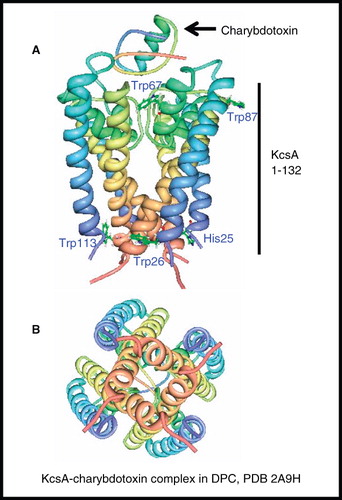
Figure 9. NMR structure of DAP12-NKG2C complex. (A) Structure of DAP12-NKG2C complex in DPC/SDS and (B) comparison of DAP12 and ζζ TM domain structures. The picture in A was produced using the PDB file and PDB Protein Workshop 3.9 (Moreland et al. Citation2005); B is adapted by permission from Macmillan Publishers Ltd: Nature Immunology (Call et al. Citation2010; copyright 2010). This Figure is reproduced in colour in Molecular Membrane Biology online.
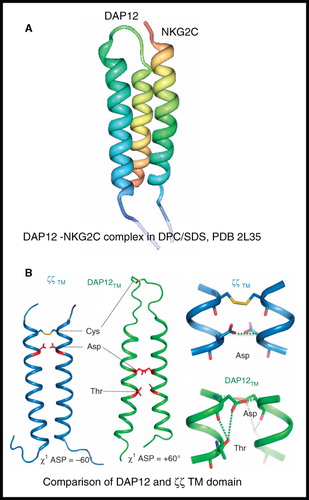
Figure 10. NMR structure of DAGK. Structure of DAGK in DPC; (A) view from the membrane plane and (B) view from the cytosol. These pictures were produced using the PDB file and PDB Protein Workshop 3.9 (Moreland et al. Citation2005). This Figure is reproduced in colour in Molecular Membrane Biology online.
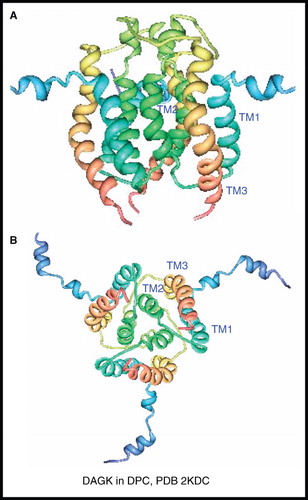
Figure 11. NMR structures of α-helical membrane proteins with four unique α-helices. Structures of (A) DsbB in DPC, (B) KdpD TM domain (397–502) in LMPG, (C) Voltage-Sensor Domain from KvaP (5–147) in D7PC and (D) nAChR β2 subunit TM domain in HFIP/H2O. All pictures were produced using the PDB file and PDB Protein Workshop 3.9 (Moreland et al. Citation2005). This Figure is reproduced in colour in Molecular Membrane Biology online.
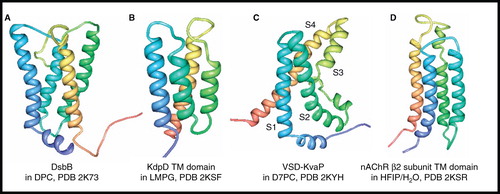
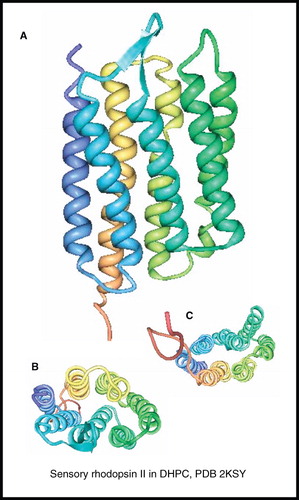
Figure 12. NMR structure of sensory rhodopsin II. Structure of sensory rhodopsin II in DHPC; (A) view from the membrane plane, (B) view from the cytosol and (C) view from the periplasm. These pictures were produced using the PDB file and PDB Protein Workshop 3.9 (Moreland et al. Citation2005). This Figure is reproduced in colour in Molecular Membrane Biology online.