Figures & data
Figure 1. Cartoon model of three membrane mimetic media. (a) A detergent micelle, (b) a two-component bicelle and (c) a lipid vesicle. The figures were generated with PyMol (W.L. DeLano, The PyMol Molecular graphics system [2002], http://www.pymol.org). This Figure is reproduced in colour in the online version of Molecular Membrane Biology.
![Figure 1. Cartoon model of three membrane mimetic media. (a) A detergent micelle, (b) a two-component bicelle and (c) a lipid vesicle. The figures were generated with PyMol (W.L. DeLano, The PyMol Molecular graphics system [2002], http://www.pymol.org). This Figure is reproduced in colour in the online version of Molecular Membrane Biology.](/cms/asset/47889384-1595-4ce6-9ba2-dbd30c93245d/imbc_a_683456_f0001_b.jpg)
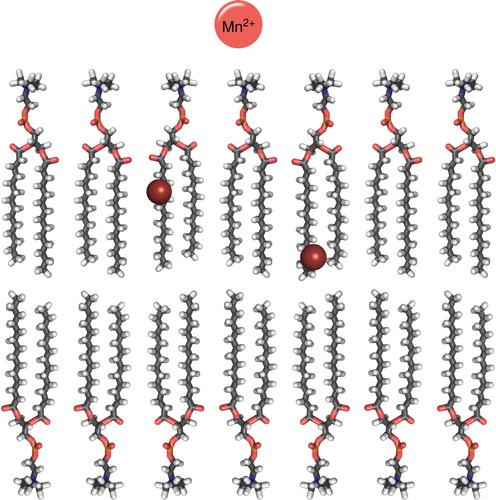
Figure 2. Molecular structures of selected phospholipids used for membrane mimetic media such as vesicles, fast-tumbling bicelles and nanodiscs.
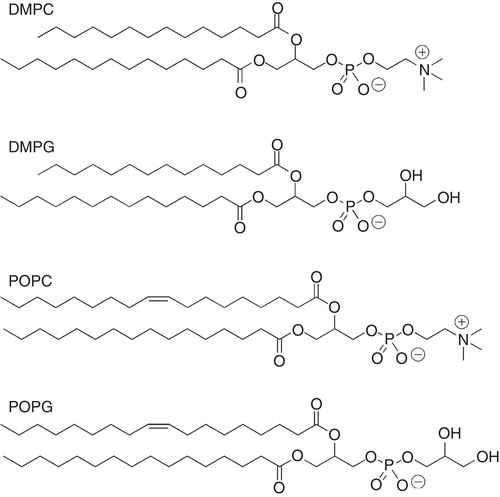
Figure 3. Molecular structure of selected detergents commonly used in solution NMR studies of peptides.
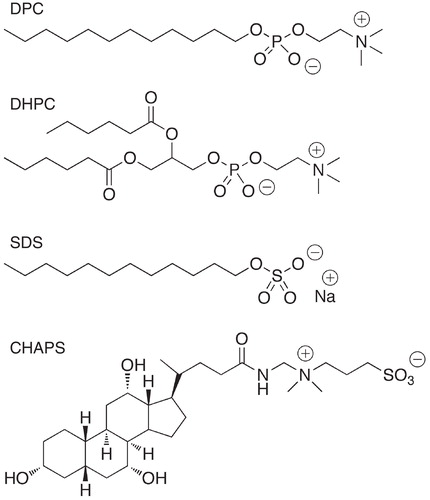
Table 1. Physical properties of some selected detergents.
Figure 4. NMR-derived solution structures of three β-barrel proteins and two helical proteins with three or more unique helices. The coordinates were taken from the PDB (www.rcsb.org). (a) OmpX in DHPC (PDB accession code 1Q9F, (b) OmpA in DPC (PDB accession code 1G90), (c) VDAC-1 in LDAO (PDB accession code 2K4T), (d) DAGK in DPC (PDB accession code 2kdc), (e) the VSD domain of KvAP in D7PC (PDB accession code 2KYH) (f) sensory rhodopsin II (residues 1–221) in D7PC (PDB accession code 2KSY), and (g) proteorhodopsin in D7PC (PDB accession code 2L6X). The figures were generated with PyMol (W.L. DeLano, The PyMol Molecular graphics system [2002], http://www.pymol.org). This Figure is reproduced in colour in the online version of Molecular Membrane Biology.
![Figure 4. NMR-derived solution structures of three β-barrel proteins and two helical proteins with three or more unique helices. The coordinates were taken from the PDB (www.rcsb.org). (a) OmpX in DHPC (PDB accession code 1Q9F, (b) OmpA in DPC (PDB accession code 1G90), (c) VDAC-1 in LDAO (PDB accession code 2K4T), (d) DAGK in DPC (PDB accession code 2kdc), (e) the VSD domain of KvAP in D7PC (PDB accession code 2KYH) (f) sensory rhodopsin II (residues 1–221) in D7PC (PDB accession code 2KSY), and (g) proteorhodopsin in D7PC (PDB accession code 2L6X). The figures were generated with PyMol (W.L. DeLano, The PyMol Molecular graphics system [2002], http://www.pymol.org). This Figure is reproduced in colour in the online version of Molecular Membrane Biology.](/cms/asset/512beafa-026c-4468-a833-c2f4726328a9/imbc_a_683456_f0004_b.jpg)
Figure 5. NMR solution structures of three cell-penetrating peptides derived in different membrane mimetic media. The structure of penetratin (a) was in q = 0.5 DMPC/DMPG/DHPC phospholipid bicelles (30% PG), of transportan (b) in q = 0.33 DMPC/DHPC, and of the N-terminal fragment (1–30) of the bovine Prion protein (c) in DHPC. The Figures were generated with PyMol (W.L. DeLano, The PyMol Molecular graphics system [2002], http://www.pymol.org). The coordinates were taken from the PDB (www.rcsb.org) (penetratin: 1OMQ, transportan: 1SMZ and bovine Prion protein-derived peptide: 1SKH). This Figure is reproduced in colour in the online version of Molecular Membrane Biology.
![Figure 5. NMR solution structures of three cell-penetrating peptides derived in different membrane mimetic media. The structure of penetratin (a) was in q = 0.5 DMPC/DMPG/DHPC phospholipid bicelles (30% PG), of transportan (b) in q = 0.33 DMPC/DHPC, and of the N-terminal fragment (1–30) of the bovine Prion protein (c) in DHPC. The Figures were generated with PyMol (W.L. DeLano, The PyMol Molecular graphics system [2002], http://www.pymol.org). The coordinates were taken from the PDB (www.rcsb.org) (penetratin: 1OMQ, transportan: 1SMZ and bovine Prion protein-derived peptide: 1SKH). This Figure is reproduced in colour in the online version of Molecular Membrane Biology.](/cms/asset/b4d32c1c-4211-4266-b910-94620f23f706/imbc_a_683456_f0005_b.jpg)
Figure 6. Mobility profile for DMPC in bicelles. 13C NOE factors for selected carbons in DMPC in q = 0.5 DMPC/CHAPS bicelles. The Figure was produced from data in (Andersson et al. Citation2007). The carbon atom labeling is indicated in the DMPC structure.
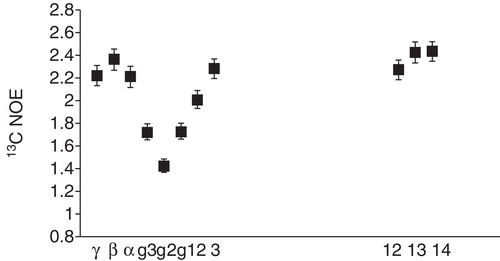
Figure 7. Cartoon over the location of commonly used spin-labeled lipids and paramagnetic metals (Mn2+) in solution NMR for elucidating peptide position in a DMPC bilayer. The spheres indicate the position the DOXYL label in 5- and 12-DOXYL labeled lipids. This Figure is reproduced in colour in the online version of Molecular Membrane Biology.