Figures & data
Figure 1. Schematic depiction of the geometrical model considered for the FRET formalism. A cylindrical membrane protein (light grey) bears a FRET donor group in an axially symmetric position (D in the right panel, which represents a side view), and is surrounded by N annular lipids (dark grey; in the left panel, which represents a top view, n = 9) in each membrane leaflet. Bulk lipids are shown with white headgroup. The donor-acceptor (A) distances to annular layer-located probes (d1 and d2, for annular lipids located at the top or bottom leaflet, respectively) are shown in the right panel, together with the exclusions distances to bulk-located probes (Re1 and Re2).
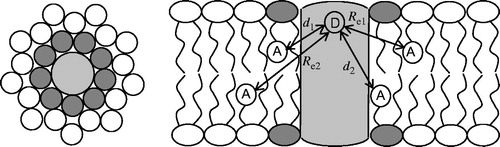
Figure 2. Structure of LacY, highlighting the FRET donor W151, relevant residue D68, and mutation site C154G. Panels A and B show side and top views, respectively.
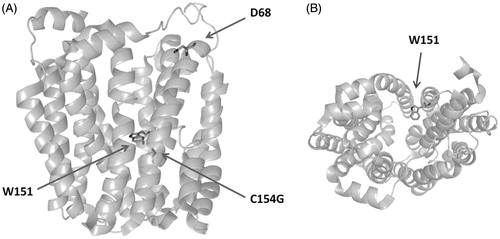
Figure 3. Structures of the pyrene-labelled lipids used as acceptors. (A) Tail-labeled lipids. R = ,
, and CH(OH)CH2OH for Pyr-PE, Pyr-PC, and Pyr-PG, respectively. (B) Head-labeled lipid, HPyr-PE.
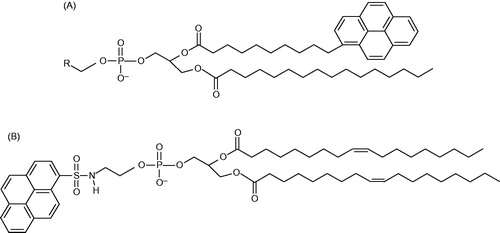
Figure 4. Comparison of experimental and theoretical values of FRET efficiency between W151 and Pyr-PG (top) and Pyr-PE (bottom) at 37 °C in POPE:POPG (3:1, mol/mol) (left), DOPE:POPG (3:1, mol/mol) (center) and DPPE:POPG (3:1, mol/mol) (right) proteoliposomes (1.5 μM LacY). Reprinted with permission from Picas et al. (Citation2010b). © 2010 Elsevier.
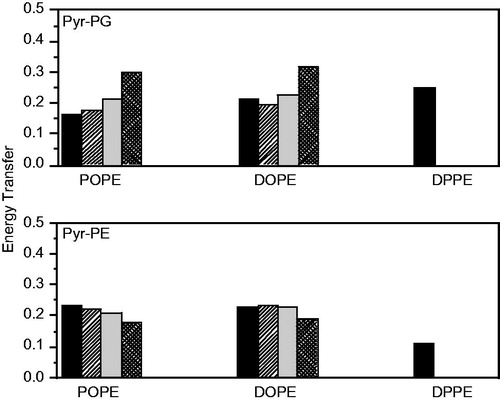
Table 1. Comparison of experimental and theoretical FRET efficiencies for ternary mixtures PE:PG:CL 67:23:10 at 37 °C. Reprinted with permission from Picas et al. (Citation2010b). © 2010 Elsevier.
Table 2. Experimental efficiencies, probabilities of each site in the annular ring of POPE or POPG vesicles being occupied by a pyrene labeled phopholipid and relative association constant toward LacY. Reprinted with permission from Suárez-Germà et al. (Citation2012a). © 2012 American Chemical Society.
Table 3. Experimental efficiencies, probabilities of each site in the annular ring of POPC or DOPC vesicles being occupied by a pyrene labeled phospholipid and relative association constant toward LacY. Reprinted with permission from Suárez-Germà et al. (Citation2012a). © 2012 American Chemical Society.
Figure 5. FRET efficiency between W151 and Pyr-PE head-labeled in different lipid matrices at 25 °C (black columns) and at 37 °C (grey columns). Proteoliposomes (1.5 μM LacY) of DOPE:POPG 3:1 (mol:mol), POPE:POPG 3:1 (mol:mol) and DPPE:POPG 3:1 (mol:mol) were doped with x = 0.0025 of label. The error bars stand for σ/√n, σ being the standard deviation and n the number of measurements performed. Reprinted with permission from Suárez-Germà et al. (Citation2013). © 2013 American Chemical Society.
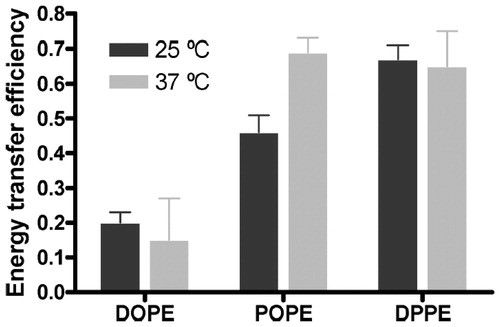
Figure 6. Comparison of normalised FRET efficiency at 37 °C between W151 of the mutant single-W C154G D68C LacY and Pyr-PE (black columns) and W151 of the single-W C154G LacY and Pyr-PE (white columns). Reprinted with permission from Suárez-Germà et al. (Citation2012b). © 2012 American Chemical Society.
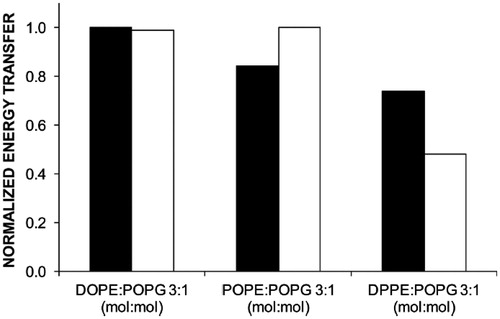
Table 4. Experimental efficiencies and probabilities µ for each site in the LacY D68C annular ring being occupied by a pyrene tail-labeled phospholipid. Reprinted with permission from Suárez-Germà et al. (Citation2012b). © 2012 American Chemical Society.
